Gene
KWMTBOMO10600
Pre Gene Modal
BGIBMGA003908
Annotation
PREDICTED:_cytochrome_P450_9e2-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.692
Sequence
CDS
ATGCGAGTGGCGCTTTCTCCGGCGTTCTCCGGAGCCCGATGTCGTAACATGGCACCGTTGATGGTGGAAAGCGCTAAGTCGGTCACAAACCACCTCCAGAAAAGAATAATTGACGAAAAGGTCATTGACATTAATAACATAACTATGAGCTACGTGAATGACGTCATAGCGTCATGCGCCTTCGGCTTCGCGGTAGACTCTCTGAAGGAACCAGACAACTGCATCTACAAGCTTGGTCAGAAAGCGATAATACAGGACACAACGCAAGTGATGAAGTTCTTTGGCTACGAGAACATTAAGACTGTTATGAAGGGGTTGAAAGTGAAAATCATCGCGACTGAAGAGGCTGAACAATTTGGTCTACTCTTCAAATCGGCCCTGAAAGCTAGAAAGGAGAAACTGGTGGGGCCGAGACCGGACTTCATACAAATATTAGTAGACGCAATGCAAGGTATGCTTATATACGAGTTAGCTAACTAG
Protein
MRVALSPAFSGARCRNMAPLMVESAKSVTNHLQKRIIDEKVIDINNITMSYVNDVIASCAFGFAVDSLKEPDNCIYKLGQKAIIQDTTQVMKFFGYENIKTVMKGLKVKIIATEEAEQFGLLFKSALKARKEKLVGPRPDFIQILVDAMQGMLIYELAN
Summary
Cofactor
heme
Similarity
Belongs to the cytochrome P450 family.
Uniprot
H9J321
A0A2A4J0W7
A0A2A4JDY3
A0A068EVM2
A0A2W1BRL3
A0A286MXM8
+ More
A0A0K8TUK8 A0A0K8TUK1 A0A212F932 E2ASC7 A0A346II56 E9JBX3 B5MF67 L0N4K3 L0N6I2 A5HKM0 H9J356 Q7YZX4 A3RIC2 A4UU25 A0A1W5ZJJ8 A0A385LJM0 A0A068EXP4 A0A3S2M2T6 B6ECI4 A0A0N1PH22 A0A385LJP1 A0A2D1QTX1 Q7YZS1 A0A194QDY1 T1PHF2 Q1KLD7 A0A195C1Q6 A0A248QFE0 A0A194RNT1 B6V7E2 A0A194R3L1 C6H0J5 E2ASC4 A0A194QDQ3 A0A286MXM7 A0A0P0EW60 A0A194R4E5 A5HMQ7 F4W701 X5D4G9 Q17HK2 A0A3S2NVP0 L0N4J7 A0A026WN38 F4W702 L0N797 B0WTQ3 E2ASC2 A0A0C5CGW0 A3RKJ5 A0A195DB16 D0FZ10 L0N725 H9J358 B0WTQ2 E9IMX5 A0A0J7KCV0 A0A0J7KCG9 A0A0J7KH63 A0A0L7L0K4 A0A195DAZ9 Q9U9C0 A0A2D1QTX7 D0VY60 A0A248QJ18 A5HKL6 A5HKL8 A4ZWB1 A0A0J7KEC8 A0A195AX20 A0A151K0Q4 A5HKL9 A0A0C5BXE6 R9Z5T9 A0A232FEF9 A0A385LJV6 B6VEL8 K7INH5 A0A232F749 A0A182GB14 A0A0L7QR58 A0A0J7K286 A0A248QEG5 Q5YJK0 A0A1V0D9E5 A0A0J7KVQ4 X5DAZ5 Q9U9C1 A0A3G5EAT6 A0A1Q3FRZ6 A0A3L8D4V8 A0A151X7X3 B7S8W6
A0A0K8TUK8 A0A0K8TUK1 A0A212F932 E2ASC7 A0A346II56 E9JBX3 B5MF67 L0N4K3 L0N6I2 A5HKM0 H9J356 Q7YZX4 A3RIC2 A4UU25 A0A1W5ZJJ8 A0A385LJM0 A0A068EXP4 A0A3S2M2T6 B6ECI4 A0A0N1PH22 A0A385LJP1 A0A2D1QTX1 Q7YZS1 A0A194QDY1 T1PHF2 Q1KLD7 A0A195C1Q6 A0A248QFE0 A0A194RNT1 B6V7E2 A0A194R3L1 C6H0J5 E2ASC4 A0A194QDQ3 A0A286MXM7 A0A0P0EW60 A0A194R4E5 A5HMQ7 F4W701 X5D4G9 Q17HK2 A0A3S2NVP0 L0N4J7 A0A026WN38 F4W702 L0N797 B0WTQ3 E2ASC2 A0A0C5CGW0 A3RKJ5 A0A195DB16 D0FZ10 L0N725 H9J358 B0WTQ2 E9IMX5 A0A0J7KCV0 A0A0J7KCG9 A0A0J7KH63 A0A0L7L0K4 A0A195DAZ9 Q9U9C0 A0A2D1QTX7 D0VY60 A0A248QJ18 A5HKL6 A5HKL8 A4ZWB1 A0A0J7KEC8 A0A195AX20 A0A151K0Q4 A5HKL9 A0A0C5BXE6 R9Z5T9 A0A232FEF9 A0A385LJV6 B6VEL8 K7INH5 A0A232F749 A0A182GB14 A0A0L7QR58 A0A0J7K286 A0A248QEG5 Q5YJK0 A0A1V0D9E5 A0A0J7KVQ4 X5DAZ5 Q9U9C1 A0A3G5EAT6 A0A1Q3FRZ6 A0A3L8D4V8 A0A151X7X3 B7S8W6
Pubmed
EMBL
BABH01037667
BABH01037668
BABH01037669
BABH01037670
BABH01037671
NWSH01003887
+ More
PCG65725.1 NWSH01001849 PCG69918.1 KM016756 AID54908.1 KZ149921 PZC77719.1 MF684346 ASX93980.1 GCVX01000063 JAI18167.1 GCVX01000068 JAI18162.1 AGBW02009651 OWR50238.1 GL442284 EFN63661.1 MH138270 AXP17211.1 GL771642 EFZ09644.1 AB381883 BAG71410.1 AK289302 BAM73829.1 AK289300 BAM73827.1 EF535808 ABQ08710.1 BABH01036639 BABH01036640 AF525031 AAP80766.1 EF415298 ABN71369.1 BABH01036638 EF488994 ABP02071.1 KY436739 ARI68319.1 MG793355 AYA28074.1 KJ671577 AID55429.1 RSAL01000054 RVE50006.1 FJ227150 ACI43222.1 KQ459169 KPJ03415.1 MG793356 AYA28075.1 KY348418 ATP15899.1 AY295775 AAP83690.1 KQ459144 KPJ03682.1 KA647545 AFP62174.1 DQ473438 ABF14737.1 KQ978344 KYM94809.1 KX443437 ASO98012.1 KQ460124 KPJ17676.1 FJ265741 ACJ04711.1 KQ460779 KPJ12393.1 FN421128 CAZ65619.1 EFN63658.1 KPJ03683.1 MF684345 ASX93979.1 KR676343 ALJ30298.1 KPJ12394.1 EF542811 ABQ18318.1 GL887802 EGI69991.1 KF701146 AHW57316.1 CH477247 EAT46155.1 RVE50007.1 AK289297 BAM73824.1 KK107147 EZA57482.1 EGI69992.1 AK289298 BAM73825.1 DS232090 EDS34511.1 EFN63656.1 KP001132 AJN91177.1 EF421989 ABO07439.1 KQ981042 KYN10061.1 AB498807 BAI47532.1 AK289299 BAM73826.1 BABH01036637 EDS34510.1 GL764255 EFZ18125.1 LBMM01009553 KMQ88036.1 KMQ88037.1 LBMM01007590 KMQ89602.1 JTDY01003828 KOB68945.1 KYN10063.1 AF172280 AAD51037.1 KY348420 ATP15901.1 AB500115 BAI49180.1 KX443436 ASO98011.1 EF535804 ABQ08706.1 EF535806 ABQ08708.2 BABH01036652 BABH01036653 EF491004 AK343209 ABP65279.1 BAM73893.1 LBMM01008897 KMQ88546.1 KQ976716 KYM76793.1 KQ981253 KYN44187.1 EF535807 ABQ08709.1 KP001128 AJN91173.1 KF056903 AGO36276.1 NNAY01000334 OXU29136.1 MG793354 AYA28073.1 FJ378716 ACJ05915.1 NNAY01000835 OXU26268.1 JXUM01009174 KQ560277 KXJ83348.1 KQ414784 KOC61112.1 LBMM01016795 KMQ84306.1 KX443439 ASO98014.1 AY390260 AAR26518.1 KY212055 ARA91619.1 LBMM01002800 KMQ94344.1 KF701145 AHW57315.1 AF172279 AAD51036.1 MH001162 AYW35335.1 GFDL01004779 JAV30266.1 QOIP01000013 RLU15532.1 KQ982431 KYQ56463.1 EF710653 ACE75341.1
PCG65725.1 NWSH01001849 PCG69918.1 KM016756 AID54908.1 KZ149921 PZC77719.1 MF684346 ASX93980.1 GCVX01000063 JAI18167.1 GCVX01000068 JAI18162.1 AGBW02009651 OWR50238.1 GL442284 EFN63661.1 MH138270 AXP17211.1 GL771642 EFZ09644.1 AB381883 BAG71410.1 AK289302 BAM73829.1 AK289300 BAM73827.1 EF535808 ABQ08710.1 BABH01036639 BABH01036640 AF525031 AAP80766.1 EF415298 ABN71369.1 BABH01036638 EF488994 ABP02071.1 KY436739 ARI68319.1 MG793355 AYA28074.1 KJ671577 AID55429.1 RSAL01000054 RVE50006.1 FJ227150 ACI43222.1 KQ459169 KPJ03415.1 MG793356 AYA28075.1 KY348418 ATP15899.1 AY295775 AAP83690.1 KQ459144 KPJ03682.1 KA647545 AFP62174.1 DQ473438 ABF14737.1 KQ978344 KYM94809.1 KX443437 ASO98012.1 KQ460124 KPJ17676.1 FJ265741 ACJ04711.1 KQ460779 KPJ12393.1 FN421128 CAZ65619.1 EFN63658.1 KPJ03683.1 MF684345 ASX93979.1 KR676343 ALJ30298.1 KPJ12394.1 EF542811 ABQ18318.1 GL887802 EGI69991.1 KF701146 AHW57316.1 CH477247 EAT46155.1 RVE50007.1 AK289297 BAM73824.1 KK107147 EZA57482.1 EGI69992.1 AK289298 BAM73825.1 DS232090 EDS34511.1 EFN63656.1 KP001132 AJN91177.1 EF421989 ABO07439.1 KQ981042 KYN10061.1 AB498807 BAI47532.1 AK289299 BAM73826.1 BABH01036637 EDS34510.1 GL764255 EFZ18125.1 LBMM01009553 KMQ88036.1 KMQ88037.1 LBMM01007590 KMQ89602.1 JTDY01003828 KOB68945.1 KYN10063.1 AF172280 AAD51037.1 KY348420 ATP15901.1 AB500115 BAI49180.1 KX443436 ASO98011.1 EF535804 ABQ08706.1 EF535806 ABQ08708.2 BABH01036652 BABH01036653 EF491004 AK343209 ABP65279.1 BAM73893.1 LBMM01008897 KMQ88546.1 KQ976716 KYM76793.1 KQ981253 KYN44187.1 EF535807 ABQ08709.1 KP001128 AJN91173.1 KF056903 AGO36276.1 NNAY01000334 OXU29136.1 MG793354 AYA28073.1 FJ378716 ACJ05915.1 NNAY01000835 OXU26268.1 JXUM01009174 KQ560277 KXJ83348.1 KQ414784 KOC61112.1 LBMM01016795 KMQ84306.1 KX443439 ASO98014.1 AY390260 AAR26518.1 KY212055 ARA91619.1 LBMM01002800 KMQ94344.1 KF701145 AHW57315.1 AF172279 AAD51036.1 MH001162 AYW35335.1 GFDL01004779 JAV30266.1 QOIP01000013 RLU15532.1 KQ982431 KYQ56463.1 EF710653 ACE75341.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000000311
UP000283053
UP000053268
+ More
UP000095301 UP000078542 UP000053240 UP000007755 UP000008820 UP000053097 UP000002320 UP000078492 UP000036403 UP000037510 UP000078540 UP000078541 UP000215335 UP000002358 UP000069940 UP000249989 UP000053825 UP000279307 UP000075809
UP000095301 UP000078542 UP000053240 UP000007755 UP000008820 UP000053097 UP000002320 UP000078492 UP000036403 UP000037510 UP000078540 UP000078541 UP000215335 UP000002358 UP000069940 UP000249989 UP000053825 UP000279307 UP000075809
PRIDE
Pfam
PF00067 p450
Interpro
SUPFAM
SSF48264
SSF48264
Gene 3D
ProteinModelPortal
H9J321
A0A2A4J0W7
A0A2A4JDY3
A0A068EVM2
A0A2W1BRL3
A0A286MXM8
+ More
A0A0K8TUK8 A0A0K8TUK1 A0A212F932 E2ASC7 A0A346II56 E9JBX3 B5MF67 L0N4K3 L0N6I2 A5HKM0 H9J356 Q7YZX4 A3RIC2 A4UU25 A0A1W5ZJJ8 A0A385LJM0 A0A068EXP4 A0A3S2M2T6 B6ECI4 A0A0N1PH22 A0A385LJP1 A0A2D1QTX1 Q7YZS1 A0A194QDY1 T1PHF2 Q1KLD7 A0A195C1Q6 A0A248QFE0 A0A194RNT1 B6V7E2 A0A194R3L1 C6H0J5 E2ASC4 A0A194QDQ3 A0A286MXM7 A0A0P0EW60 A0A194R4E5 A5HMQ7 F4W701 X5D4G9 Q17HK2 A0A3S2NVP0 L0N4J7 A0A026WN38 F4W702 L0N797 B0WTQ3 E2ASC2 A0A0C5CGW0 A3RKJ5 A0A195DB16 D0FZ10 L0N725 H9J358 B0WTQ2 E9IMX5 A0A0J7KCV0 A0A0J7KCG9 A0A0J7KH63 A0A0L7L0K4 A0A195DAZ9 Q9U9C0 A0A2D1QTX7 D0VY60 A0A248QJ18 A5HKL6 A5HKL8 A4ZWB1 A0A0J7KEC8 A0A195AX20 A0A151K0Q4 A5HKL9 A0A0C5BXE6 R9Z5T9 A0A232FEF9 A0A385LJV6 B6VEL8 K7INH5 A0A232F749 A0A182GB14 A0A0L7QR58 A0A0J7K286 A0A248QEG5 Q5YJK0 A0A1V0D9E5 A0A0J7KVQ4 X5DAZ5 Q9U9C1 A0A3G5EAT6 A0A1Q3FRZ6 A0A3L8D4V8 A0A151X7X3 B7S8W6
A0A0K8TUK8 A0A0K8TUK1 A0A212F932 E2ASC7 A0A346II56 E9JBX3 B5MF67 L0N4K3 L0N6I2 A5HKM0 H9J356 Q7YZX4 A3RIC2 A4UU25 A0A1W5ZJJ8 A0A385LJM0 A0A068EXP4 A0A3S2M2T6 B6ECI4 A0A0N1PH22 A0A385LJP1 A0A2D1QTX1 Q7YZS1 A0A194QDY1 T1PHF2 Q1KLD7 A0A195C1Q6 A0A248QFE0 A0A194RNT1 B6V7E2 A0A194R3L1 C6H0J5 E2ASC4 A0A194QDQ3 A0A286MXM7 A0A0P0EW60 A0A194R4E5 A5HMQ7 F4W701 X5D4G9 Q17HK2 A0A3S2NVP0 L0N4J7 A0A026WN38 F4W702 L0N797 B0WTQ3 E2ASC2 A0A0C5CGW0 A3RKJ5 A0A195DB16 D0FZ10 L0N725 H9J358 B0WTQ2 E9IMX5 A0A0J7KCV0 A0A0J7KCG9 A0A0J7KH63 A0A0L7L0K4 A0A195DAZ9 Q9U9C0 A0A2D1QTX7 D0VY60 A0A248QJ18 A5HKL6 A5HKL8 A4ZWB1 A0A0J7KEC8 A0A195AX20 A0A151K0Q4 A5HKL9 A0A0C5BXE6 R9Z5T9 A0A232FEF9 A0A385LJV6 B6VEL8 K7INH5 A0A232F749 A0A182GB14 A0A0L7QR58 A0A0J7K286 A0A248QEG5 Q5YJK0 A0A1V0D9E5 A0A0J7KVQ4 X5DAZ5 Q9U9C1 A0A3G5EAT6 A0A1Q3FRZ6 A0A3L8D4V8 A0A151X7X3 B7S8W6
PDB
6MA7
E-value=3.44981e-07,
Score=124
Ontologies
GO
Topology
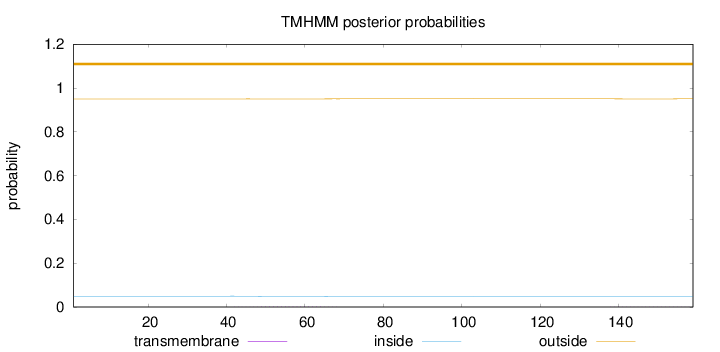
Length:
159
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0403
Exp number, first 60 AAs:
0.02232
Total prob of N-in:
0.04956
outside
1 - 159
Population Genetic Test Statistics
Pi
172.96113
Theta
154.876675
Tajima's D
-0.719549
CLR
2.89083
CSRT
0.191990400479976
Interpretation
Uncertain
