Pre Gene Modal
BGIBMGA003910
Annotation
PREDICTED:_pyridoxal-dependent_decarboxylase_domain-containing_protein_1_isoform_X2_[Bombyx_mori]
Full name
Pyridoxal-dependent decarboxylase domain-containing protein 1
Location in the cell
Cytoplasmic Reliability : 1.611 Nuclear Reliability : 1.745
Sequence
CDS
ATGGGAGATGCATCAGCTTCGGACACCGAGTCCAATAAACCTAGTCCTATCAACGAACCACATTTAGAACTGGAGCGGCGTCCCTTCGGTGGGCTCGAGTTTCAAGTATCTGAAGTAGTAGAAAGGCTCGAAGCCGGGGTTAATGCACAGGATGCTGCCGATGACGAGAAAAAGCGTGAACCGCGTAAAATCAGTACCGGCTTTTTTGAACCCGAAGAAACACCAATGGATGAAATTCTGAAAATTTTAGAGGAACTAGTGATCAAGACCGACCCTAGTTGCGAGAGCTTAGAGCCTCCGCTGTTGCCGACGGATGCGGTGACTCGTGCGGCCATTCTTTCTCATAGTATTTCAGCATTATTTGGTCGCTTAGAAAGAAGTCACGCAGCGCGCCTCGGTACTCACATAGCAAGCGAAACGACTCGATGGATGGCTCACATGTTTAGGCTATCAGACTACAATGCATATTATCATCAAGAACAGCTTGAAGGTCTTGTGCGGGTCACAAGGATGCTGCTCCATCACAGATACCCTAGATATTTAGAAGATGGAGCATTGGCGTTTGCAAATCGTCTACCCTGCATATACAGTTGCGTTGCGAGTCCACTGGGGATCGTTCAGCACCTCTGCAGACAGCTGGGCTTACCGCTGGCTTGTGTTAGGCCAGTTCCAGTAAATCCTTCAGGTCGAGGAATGGACATAGACGCGTTAGAGAGGCTATGTGACGAGGACGTCGCCGCCAATCGCACCCCGCTGCTGGTGCTGGGAGAAGTGGGAGCTCCGCCCCTAGGTTGGGGCACTCCGCTGTACACCCTTGGTGAAATCTGCGCTAGAAGAAACCTGCACCTACACGTCAGAGGGCACGCGTTAGCCTTACCTGGTGCCAGCCTGAAAGAGCAGACTTACAGTATACCGGACTCGTTGACGCTGACTCCAGGACCGTGGTTCGGCGTGCCGGGCCTGCCTACTGTCACGTTTTACAAAATTCCAGAATCGGTAACAGCCAACGATCAATCTAAGGGCGTTAACGCGGTAAGTGAAGAATATTCAATTGTACAATAG
Protein
MGDASASDTESNKPSPINEPHLELERRPFGGLEFQVSEVVERLEAGVNAQDAADDEKKREPRKISTGFFEPEETPMDEILKILEELVIKTDPSCESLEPPLLPTDAVTRAAILSHSISALFGRLERSHAARLGTHIASETTRWMAHMFRLSDYNAYYHQEQLEGLVRVTRMLLHHRYPRYLEDGALAFANRLPCIYSCVASPLGIVQHLCRQLGLPLACVRPVPVNPSGRGMDIDALERLCDEDVAANRTPLLVLGEVGAPPLGWGTPLYTLGEICARRNLHLHVRGHALALPGASLKEQTYSIPDSLTLTPGPWFGVPGLPTVTFYKIPESVTANDQSKGVNAVSEEYSIVQ
Summary
Cofactor
pyridoxal 5'-phosphate
Similarity
Belongs to the group II decarboxylase family.
Keywords
Decarboxylase
Lyase
Pyridoxal phosphate
Feature
chain Pyridoxal-dependent decarboxylase domain-containing protein 1
Uniprot
A0A2H1VBZ8
A0A2A4J667
A0A212EL63
A0A2W1BRM4
H9J323
A0A1E1VZP7
+ More
A0A194PPV2 A0A0N1PGF8 A0A2Z5U729 A0A067RWL3 A0A2J7PLV7 A0A2P8Z2L2 A0A1Y1JUJ8 D6WYJ0 A0A0T6AV11 A0A1B6CKL1 U4USC4 E2BTH1 A0A087ZQM4 A0A154P8L0 A0A2A3E3X9 F4WTZ8 A0A158P110 A0A151J1V8 A0A0L7QPP3 A0A1B6F8Q9 E2AXN4 N6TC82 A0A310SS32 A0A151WWY6 A0A195AUQ0 A0A1B6HIF9 A0A0M9A8V4 A0A232EYQ3 A0A3L8E2A5 A0A0C9QZX3 K7IZ82 A0A0V0G4W6 A0A069DWD2 E0VSR3 A0A0J7KNC7 A0A224X8N3 A0A146LA38 A0A146MII3 A0A026WER0 A0A1W5AJ52 A0A1W5AIL0 A0A1W5AHF4 A0A1W5AAM3 A0A2G8L6F9 A0A1L8EXK7 Q6DF78 A0A1S3K002 F7BG81 F6R560 A0A1B0C9Q3 R7UV22 A0A1L8EQ88 A0A1L8E297 A0A2G8JCI3 A0A1L8E2C1 A0A091VF94 A0A093I442 A0A0B8RXL8 U3FX57 B0W7U7 A0A2D4KCK1 H9G850 A0A1V4KCN6 A0A1U7RXI7 G3WAL5 A0A151N1D4 A0A2I0LRE4 U3IC71 A0A2D4LB04 A0A094KJB4 A0A091PZP3 A0A093HSP3 A0A091P907 A0A091HEZ4 A0A3B3QA03 A0A091KW96 A0A1I8P2F7
A0A194PPV2 A0A0N1PGF8 A0A2Z5U729 A0A067RWL3 A0A2J7PLV7 A0A2P8Z2L2 A0A1Y1JUJ8 D6WYJ0 A0A0T6AV11 A0A1B6CKL1 U4USC4 E2BTH1 A0A087ZQM4 A0A154P8L0 A0A2A3E3X9 F4WTZ8 A0A158P110 A0A151J1V8 A0A0L7QPP3 A0A1B6F8Q9 E2AXN4 N6TC82 A0A310SS32 A0A151WWY6 A0A195AUQ0 A0A1B6HIF9 A0A0M9A8V4 A0A232EYQ3 A0A3L8E2A5 A0A0C9QZX3 K7IZ82 A0A0V0G4W6 A0A069DWD2 E0VSR3 A0A0J7KNC7 A0A224X8N3 A0A146LA38 A0A146MII3 A0A026WER0 A0A1W5AJ52 A0A1W5AIL0 A0A1W5AHF4 A0A1W5AAM3 A0A2G8L6F9 A0A1L8EXK7 Q6DF78 A0A1S3K002 F7BG81 F6R560 A0A1B0C9Q3 R7UV22 A0A1L8EQ88 A0A1L8E297 A0A2G8JCI3 A0A1L8E2C1 A0A091VF94 A0A093I442 A0A0B8RXL8 U3FX57 B0W7U7 A0A2D4KCK1 H9G850 A0A1V4KCN6 A0A1U7RXI7 G3WAL5 A0A151N1D4 A0A2I0LRE4 U3IC71 A0A2D4LB04 A0A094KJB4 A0A091PZP3 A0A093HSP3 A0A091P907 A0A091HEZ4 A0A3B3QA03 A0A091KW96 A0A1I8P2F7
EC Number
4.1.1.-
Pubmed
22118469
28756777
19121390
26354079
24845553
29403074
+ More
28004739 18362917 19820115 23537049 20798317 21719571 21347285 28648823 30249741 20075255 26334808 20566863 26823975 24508170 29023486 27762356 20431018 17495919 23254933 25476704 23915248 21881562 21709235 22293439 23371554 23749191 29240929
28004739 18362917 19820115 23537049 20798317 21719571 21347285 28648823 30249741 20075255 26334808 20566863 26823975 24508170 29023486 27762356 20431018 17495919 23254933 25476704 23915248 21881562 21709235 22293439 23371554 23749191 29240929
EMBL
ODYU01001747
SOQ38369.1
NWSH01002881
PCG67379.1
AGBW02014130
OWR42225.1
+ More
KZ149921 PZC77742.1 BABH01037661 BABH01037662 BABH01037663 BABH01037664 GDQN01010892 JAT80162.1 KQ459603 KPI93160.1 KQ458497 KPJ05861.1 AP017481 BBB06729.1 KK852421 KDR24299.1 NEVH01024423 PNF17324.1 PYGN01000224 PSN50728.1 GEZM01103195 JAV51470.1 KQ971362 EFA07871.1 LJIG01022754 KRT78898.1 GEDC01024235 GEDC01023279 JAS13063.1 JAS14019.1 KB632428 ERL95418.1 GL450410 EFN80999.1 KQ434846 KZC08255.1 KZ288386 PBC26463.1 GL888345 EGI62355.1 ADTU01005935 KQ980514 KYN15707.1 KQ414806 KOC60600.1 GECZ01023209 JAS46560.1 GL443604 EFN61815.1 APGK01035703 KB740928 ENN77914.1 KQ761005 OAD58404.1 KQ982691 KYQ52175.1 KQ976738 KYM75787.1 GECU01033206 JAS74500.1 KQ435724 KOX78313.1 NNAY01001571 OXU23562.1 QOIP01000001 RLU26329.1 GBYB01009354 GBYB01009359 JAG79121.1 JAG79126.1 GECL01003038 JAP03086.1 GBGD01000669 JAC88220.1 DS235755 EEB16419.1 LBMM01005099 KMQ91817.1 GFTR01007740 JAW08686.1 GDHC01013431 JAQ05198.1 GDHC01019724 GDHC01000083 JAP98904.1 JAQ18546.1 KK107242 EZA54552.1 MRZV01000198 PIK55859.1 CM004482 OCT64076.1 BC076864 AAMC01072836 AJWK01002591 AMQN01006037 KB297448 ELU10488.1 CM004483 OCT61481.1 GFDF01001224 JAV12860.1 MRZV01002549 PIK33455.1 GFDF01001223 JAV12861.1 KK428118 KFQ88303.1 KL214745 KFV61541.1 GBSH01002148 JAG66878.1 GAEP01000445 JAB54376.1 DS231855 EDS38241.1 IACL01047663 LAB06423.1 AAWZ02030569 AAWZ02030570 LSYS01003774 OPJ82230.1 AEFK01077636 AEFK01077637 AEFK01077638 AEFK01077639 AEFK01077640 AEFK01077641 AKHW03004154 KYO30558.1 AKCR02000124 PKK19997.1 ADON01065060 IACM01008690 LAB18078.1 KL350657 KFZ58685.1 KK683030 KFQ13417.1 KL206788 KFV85708.1 KK655175 KFQ04407.1 KL532858 KFO93397.1 KK757218 KFP43748.1
KZ149921 PZC77742.1 BABH01037661 BABH01037662 BABH01037663 BABH01037664 GDQN01010892 JAT80162.1 KQ459603 KPI93160.1 KQ458497 KPJ05861.1 AP017481 BBB06729.1 KK852421 KDR24299.1 NEVH01024423 PNF17324.1 PYGN01000224 PSN50728.1 GEZM01103195 JAV51470.1 KQ971362 EFA07871.1 LJIG01022754 KRT78898.1 GEDC01024235 GEDC01023279 JAS13063.1 JAS14019.1 KB632428 ERL95418.1 GL450410 EFN80999.1 KQ434846 KZC08255.1 KZ288386 PBC26463.1 GL888345 EGI62355.1 ADTU01005935 KQ980514 KYN15707.1 KQ414806 KOC60600.1 GECZ01023209 JAS46560.1 GL443604 EFN61815.1 APGK01035703 KB740928 ENN77914.1 KQ761005 OAD58404.1 KQ982691 KYQ52175.1 KQ976738 KYM75787.1 GECU01033206 JAS74500.1 KQ435724 KOX78313.1 NNAY01001571 OXU23562.1 QOIP01000001 RLU26329.1 GBYB01009354 GBYB01009359 JAG79121.1 JAG79126.1 GECL01003038 JAP03086.1 GBGD01000669 JAC88220.1 DS235755 EEB16419.1 LBMM01005099 KMQ91817.1 GFTR01007740 JAW08686.1 GDHC01013431 JAQ05198.1 GDHC01019724 GDHC01000083 JAP98904.1 JAQ18546.1 KK107242 EZA54552.1 MRZV01000198 PIK55859.1 CM004482 OCT64076.1 BC076864 AAMC01072836 AJWK01002591 AMQN01006037 KB297448 ELU10488.1 CM004483 OCT61481.1 GFDF01001224 JAV12860.1 MRZV01002549 PIK33455.1 GFDF01001223 JAV12861.1 KK428118 KFQ88303.1 KL214745 KFV61541.1 GBSH01002148 JAG66878.1 GAEP01000445 JAB54376.1 DS231855 EDS38241.1 IACL01047663 LAB06423.1 AAWZ02030569 AAWZ02030570 LSYS01003774 OPJ82230.1 AEFK01077636 AEFK01077637 AEFK01077638 AEFK01077639 AEFK01077640 AEFK01077641 AKHW03004154 KYO30558.1 AKCR02000124 PKK19997.1 ADON01065060 IACM01008690 LAB18078.1 KL350657 KFZ58685.1 KK683030 KFQ13417.1 KL206788 KFV85708.1 KK655175 KFQ04407.1 KL532858 KFO93397.1 KK757218 KFP43748.1
Proteomes
UP000218220
UP000007151
UP000005204
UP000053268
UP000027135
UP000235965
+ More
UP000245037 UP000007266 UP000030742 UP000008237 UP000005203 UP000076502 UP000242457 UP000007755 UP000005205 UP000078492 UP000053825 UP000000311 UP000019118 UP000075809 UP000078540 UP000053105 UP000215335 UP000279307 UP000002358 UP000009046 UP000036403 UP000053097 UP000192224 UP000230750 UP000186698 UP000085678 UP000008143 UP000002280 UP000092461 UP000014760 UP000053875 UP000002320 UP000001646 UP000190648 UP000189705 UP000007648 UP000050525 UP000053872 UP000016666 UP000053584 UP000261540 UP000095300
UP000245037 UP000007266 UP000030742 UP000008237 UP000005203 UP000076502 UP000242457 UP000007755 UP000005205 UP000078492 UP000053825 UP000000311 UP000019118 UP000075809 UP000078540 UP000053105 UP000215335 UP000279307 UP000002358 UP000009046 UP000036403 UP000053097 UP000192224 UP000230750 UP000186698 UP000085678 UP000008143 UP000002280 UP000092461 UP000014760 UP000053875 UP000002320 UP000001646 UP000190648 UP000189705 UP000007648 UP000050525 UP000053872 UP000016666 UP000053584 UP000261540 UP000095300
Interpro
SUPFAM
SSF53383
SSF53383
Gene 3D
ProteinModelPortal
A0A2H1VBZ8
A0A2A4J667
A0A212EL63
A0A2W1BRM4
H9J323
A0A1E1VZP7
+ More
A0A194PPV2 A0A0N1PGF8 A0A2Z5U729 A0A067RWL3 A0A2J7PLV7 A0A2P8Z2L2 A0A1Y1JUJ8 D6WYJ0 A0A0T6AV11 A0A1B6CKL1 U4USC4 E2BTH1 A0A087ZQM4 A0A154P8L0 A0A2A3E3X9 F4WTZ8 A0A158P110 A0A151J1V8 A0A0L7QPP3 A0A1B6F8Q9 E2AXN4 N6TC82 A0A310SS32 A0A151WWY6 A0A195AUQ0 A0A1B6HIF9 A0A0M9A8V4 A0A232EYQ3 A0A3L8E2A5 A0A0C9QZX3 K7IZ82 A0A0V0G4W6 A0A069DWD2 E0VSR3 A0A0J7KNC7 A0A224X8N3 A0A146LA38 A0A146MII3 A0A026WER0 A0A1W5AJ52 A0A1W5AIL0 A0A1W5AHF4 A0A1W5AAM3 A0A2G8L6F9 A0A1L8EXK7 Q6DF78 A0A1S3K002 F7BG81 F6R560 A0A1B0C9Q3 R7UV22 A0A1L8EQ88 A0A1L8E297 A0A2G8JCI3 A0A1L8E2C1 A0A091VF94 A0A093I442 A0A0B8RXL8 U3FX57 B0W7U7 A0A2D4KCK1 H9G850 A0A1V4KCN6 A0A1U7RXI7 G3WAL5 A0A151N1D4 A0A2I0LRE4 U3IC71 A0A2D4LB04 A0A094KJB4 A0A091PZP3 A0A093HSP3 A0A091P907 A0A091HEZ4 A0A3B3QA03 A0A091KW96 A0A1I8P2F7
A0A194PPV2 A0A0N1PGF8 A0A2Z5U729 A0A067RWL3 A0A2J7PLV7 A0A2P8Z2L2 A0A1Y1JUJ8 D6WYJ0 A0A0T6AV11 A0A1B6CKL1 U4USC4 E2BTH1 A0A087ZQM4 A0A154P8L0 A0A2A3E3X9 F4WTZ8 A0A158P110 A0A151J1V8 A0A0L7QPP3 A0A1B6F8Q9 E2AXN4 N6TC82 A0A310SS32 A0A151WWY6 A0A195AUQ0 A0A1B6HIF9 A0A0M9A8V4 A0A232EYQ3 A0A3L8E2A5 A0A0C9QZX3 K7IZ82 A0A0V0G4W6 A0A069DWD2 E0VSR3 A0A0J7KNC7 A0A224X8N3 A0A146LA38 A0A146MII3 A0A026WER0 A0A1W5AJ52 A0A1W5AIL0 A0A1W5AHF4 A0A1W5AAM3 A0A2G8L6F9 A0A1L8EXK7 Q6DF78 A0A1S3K002 F7BG81 F6R560 A0A1B0C9Q3 R7UV22 A0A1L8EQ88 A0A1L8E297 A0A2G8JCI3 A0A1L8E2C1 A0A091VF94 A0A093I442 A0A0B8RXL8 U3FX57 B0W7U7 A0A2D4KCK1 H9G850 A0A1V4KCN6 A0A1U7RXI7 G3WAL5 A0A151N1D4 A0A2I0LRE4 U3IC71 A0A2D4LB04 A0A094KJB4 A0A091PZP3 A0A093HSP3 A0A091P907 A0A091HEZ4 A0A3B3QA03 A0A091KW96 A0A1I8P2F7
PDB
4E1O
E-value=0.00694892,
Score=92
Ontologies
KEGG
GO
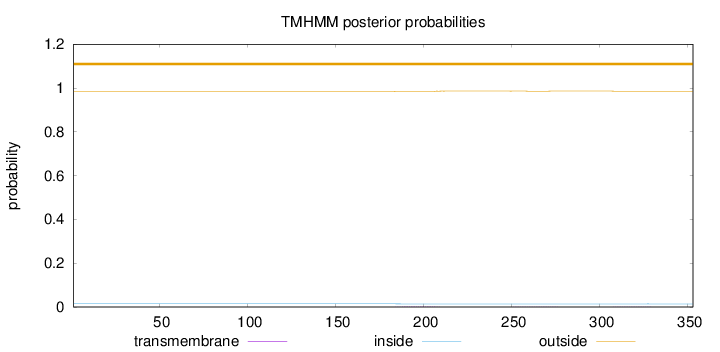
Topology
Length:
353
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0457700000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01491
outside
1 - 353
Population Genetic Test Statistics
Pi
173.400777
Theta
154.808678
Tajima's D
0.159694
CLR
102.302978
CSRT
0.415379231038448
Interpretation
Uncertain
