Gene
KWMTBOMO10591 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003923
Annotation
ribosomal_protein_S24_[Bombyx_mori]
Full name
40S ribosomal protein S24
Location in the cell
Cytoplasmic Reliability : 2.035 Mitochondrial Reliability : 1.339
Sequence
CDS
ATGAGTGAAGGAACAGCGACTATTCGCACTCGCAAGTTCATGACCAACAGATTGTTGGCGCGCAAGCAGATGGTTTGCGATGTTTTACATCCAGGAAAACCAACCGTCAGCAAGACCGAGATCCGTGAGAAGCTCGCCAAAATGTACAAGGTTACTCCCGATGTAGTGTTCGTATTCGGTTTCAAGACAAACTTCGGAGGTGGCAAGTCAACTGGATTCGCTTTGATCTACGACACACTCGATCTGGCCAAGAAGTTCGAGCCCAAGCACAGGTTAGCCCGCCACGGCCTGTACGAGAAGAAGAGGCCCACGCGCAAACAGCGTAAAGAACGTAAGAACAGAATGAAGAAGGTCCGCGGTACCAAGAAATCTAAAGTAGGTGCGGCCTCCAAGAAGTGA
Protein
MSEGTATIRTRKFMTNRLLARKQMVCDVLHPGKPTVSKTEIREKLAKMYKVTPDVVFVFGFKTNFGGGKSTGFALIYDTLDLAKKFEPKHRLARHGLYEKKRPTRKQRKERKNRMKKVRGTKKSKVGAASKK
Summary
Similarity
Belongs to the eukaryotic ribosomal protein eS24 family.
Keywords
Ribonucleoprotein
Ribosomal protein
Feature
chain 40S ribosomal protein S24
Uniprot
I4DMD8
A0A194RK09
D1LYK8
A0A212EL84
Q6PUF7
I4DIS6
+ More
A7KCX7 G0ZED1 A0A3S2TDB1 A0A0L7K9A9 E3UKP3 A0A2W1BVS8 A0A2A4IY20 A0A0K8TH14 A0A2H1VC29 Q962Q6 S4PV71 E7DZ12 Q6F461 Q4LB02 Q4LB01 V5I9Q1 D1M7Z3 A0A158NYH2 E2A641 A0A0J7LA87 A0A151J9M0 D6WFK5 A0A195CVW2 E2C1X5 F4WT28 E9JCY5 Q4LB03 A0A195F7B6 A0A026WMG3 Q4LAZ9 A0A195AZ84 A0A1E1WM94 N6UCH0 J3JWJ3 A0A088A5I5 V9IDW0 A0A0C9QHN1 A0A0C9QGY4 A0A1B6JX74 A0A171B9G5 A0A151WHJ6 A0A023F9E9 A0A224XZS8 K7IZZ4 A0A2A3EGV2 A0A109NIA6 A0A1D1XMT2 A0A0N0U5F1 A0A067QME5 A0A310SLI5 R4FPG8 A5HMP1 A0A0A9Z7E1 A0A0K8SQ58 A0A0P4VM75 A0A1B6DK47 A0A1W5I1D7 A0A069DNQ9 A0A1B6DKS0 A0A1B6D2S6 A0A154P6P6 A0A0L7QTD8 A0A2P8ZDU5 A0A2R7WZR0 A0A1B6K3J5 Q56FE4 A0A1W4VDA6 B3MEC3 A0A1J1HE02 A0A3B0JNX3 B4GC23 Q290H1 A0A0K8TQ75 A0A1B0D3Q4 A0A182K138 B4I872 B3NLI3 B4QHJ7 Q9W229 Q6XIP3 W8BKK8 A0A182N803 B4J4H7 D3TPY6 A0A1A9XIY8 A0A1A9WNJ3 B4MQG1 A0A0M4E0V6 A0A1A9VKQ7 A0A1A9ZKW2 A0A084WLA6 A0A1L8DS36 A0A1Q3F976 B0WPH5
A7KCX7 G0ZED1 A0A3S2TDB1 A0A0L7K9A9 E3UKP3 A0A2W1BVS8 A0A2A4IY20 A0A0K8TH14 A0A2H1VC29 Q962Q6 S4PV71 E7DZ12 Q6F461 Q4LB02 Q4LB01 V5I9Q1 D1M7Z3 A0A158NYH2 E2A641 A0A0J7LA87 A0A151J9M0 D6WFK5 A0A195CVW2 E2C1X5 F4WT28 E9JCY5 Q4LB03 A0A195F7B6 A0A026WMG3 Q4LAZ9 A0A195AZ84 A0A1E1WM94 N6UCH0 J3JWJ3 A0A088A5I5 V9IDW0 A0A0C9QHN1 A0A0C9QGY4 A0A1B6JX74 A0A171B9G5 A0A151WHJ6 A0A023F9E9 A0A224XZS8 K7IZZ4 A0A2A3EGV2 A0A109NIA6 A0A1D1XMT2 A0A0N0U5F1 A0A067QME5 A0A310SLI5 R4FPG8 A5HMP1 A0A0A9Z7E1 A0A0K8SQ58 A0A0P4VM75 A0A1B6DK47 A0A1W5I1D7 A0A069DNQ9 A0A1B6DKS0 A0A1B6D2S6 A0A154P6P6 A0A0L7QTD8 A0A2P8ZDU5 A0A2R7WZR0 A0A1B6K3J5 Q56FE4 A0A1W4VDA6 B3MEC3 A0A1J1HE02 A0A3B0JNX3 B4GC23 Q290H1 A0A0K8TQ75 A0A1B0D3Q4 A0A182K138 B4I872 B3NLI3 B4QHJ7 Q9W229 Q6XIP3 W8BKK8 A0A182N803 B4J4H7 D3TPY6 A0A1A9XIY8 A0A1A9WNJ3 B4MQG1 A0A0M4E0V6 A0A1A9VKQ7 A0A1A9ZKW2 A0A084WLA6 A0A1L8DS36 A0A1Q3F976 B0WPH5
Pubmed
22651552
26354079
22118469
19121390
17603110
26227816
+ More
28756777 14668217 23622113 21347285 20798317 18362917 19820115 21719571 21282665 24508170 23537049 22516182 25474469 20075255 24845553 25401762 26823975 27129103 26334808 29403074 17994087 15632085 23185243 26369729 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 23636399 26109357 26109356 14525923 17550304 24495485 20353571 24438588
28756777 14668217 23622113 21347285 20798317 18362917 19820115 21719571 21282665 24508170 23537049 22516182 25474469 20075255 24845553 25401762 26823975 27129103 26334808 29403074 17994087 15632085 23185243 26369729 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 23636399 26109357 26109356 14525923 17550304 24495485 20353571 24438588
EMBL
AK402456
BAM19078.1
KQ460124
KPJ17670.1
GU084282
ACY95315.1
+ More
AGBW02014130 OWR42227.1 BABH01037652 AY578155 AY769339 AAS91555.1 AAV34881.1 AK401194 KQ459603 BAM17816.1 KPI93157.1 EF207970 ABS57445.1 JF265009 AEL28840.1 RSAL01000304 RVE42719.1 JTDY01010218 KOB59867.1 HM449917 ADO33052.1 KZ149921 PZC77734.1 NWSH01005491 PCG64164.1 GBRD01000934 JAG64887.1 ODYU01001747 SOQ38365.1 AF400220 GAIX01009048 JAA83512.1 HQ424715 ADT80659.1 AB180429 BAD26673.1 AM040000 CAJ01865.1 AM040001 CAJ01866.1 GALX01002545 JAB65921.1 GU120417 ACY71267.1 ADTU01003827 GL437108 EFN71100.1 LBMM01000066 KMR05101.1 KQ979403 KYN21717.1 KQ971328 EFA00935.2 KQ977220 KYN04823.1 GL452008 EFN78087.1 GL888331 EGI62641.1 GL771866 EFZ09298.1 AM039999 CAJ01864.1 KQ981768 KYN35964.1 KK107151 EZA57262.1 AM040003 CAJ01868.1 KQ976701 KYM77269.1 GDQN01003078 JAT87976.1 APGK01034467 KB740914 KB631869 ENN78331.1 ERL86839.1 BT127611 AEE62573.1 JR040814 JR040815 AEY59313.1 AEY59314.1 GBYB01003039 JAG72806.1 GBYB01002739 JAG72506.1 GECU01003902 JAT03805.1 GEMB01000108 JAS03003.1 KQ983114 KYQ47311.1 GBBI01000590 JAC18122.1 GFTR01002304 JAW14122.1 KZ288262 PBC30386.1 KC507320 AHB12443.1 GDJX01024239 JAT43697.1 KQ435775 KOX75024.1 KK853170 KDR10346.1 KQ761592 OAD57369.1 GAHY01000889 JAA76621.1 EF541357 ABQ18248.1 GBHO01004326 GBRD01010559 GDHC01002003 JAG39278.1 JAG55265.1 JAQ16626.1 GBRD01010560 JAG55264.1 GDKW01003258 JAI53337.1 GEDC01011229 JAS26069.1 KJ650197 AHY30857.1 GBGD01003439 JAC85450.1 GEDC01011019 JAS26279.1 GEDC01017311 JAS19987.1 KQ434817 KZC06868.1 KQ414747 KOC61880.1 PYGN01000086 PSN54665.1 KK855850 PTY24115.1 GECU01001675 JAT06032.1 AY961556 AAX62458.1 CH902619 EDV36529.1 CVRI01000001 CRK86221.1 OUUW01000001 SPP74341.1 CH479181 EDW32366.1 CM000071 EAL25391.1 KRT02150.1 GDAI01001305 JAI16298.1 AJVK01011006 CH480824 EDW56797.1 CH954179 EDV54899.1 CM000362 CM002911 EDX08226.1 KMY95821.1 AE013599 AY113512 AAF46871.1 AAM29517.1 AY231786 AY232085 CM000158 AAR09809.1 EDW91812.1 GAMC01004690 JAC01866.1 CH916367 EDW01659.1 EZ423488 ADD19764.1 CH963849 EDW74350.1 CP012523 ALC38866.1 ATLV01024210 KE525350 KFB51000.1 GFDF01004949 JAV09135.1 GFDL01010953 JAV24092.1 DS232024 EDS32363.1
AGBW02014130 OWR42227.1 BABH01037652 AY578155 AY769339 AAS91555.1 AAV34881.1 AK401194 KQ459603 BAM17816.1 KPI93157.1 EF207970 ABS57445.1 JF265009 AEL28840.1 RSAL01000304 RVE42719.1 JTDY01010218 KOB59867.1 HM449917 ADO33052.1 KZ149921 PZC77734.1 NWSH01005491 PCG64164.1 GBRD01000934 JAG64887.1 ODYU01001747 SOQ38365.1 AF400220 GAIX01009048 JAA83512.1 HQ424715 ADT80659.1 AB180429 BAD26673.1 AM040000 CAJ01865.1 AM040001 CAJ01866.1 GALX01002545 JAB65921.1 GU120417 ACY71267.1 ADTU01003827 GL437108 EFN71100.1 LBMM01000066 KMR05101.1 KQ979403 KYN21717.1 KQ971328 EFA00935.2 KQ977220 KYN04823.1 GL452008 EFN78087.1 GL888331 EGI62641.1 GL771866 EFZ09298.1 AM039999 CAJ01864.1 KQ981768 KYN35964.1 KK107151 EZA57262.1 AM040003 CAJ01868.1 KQ976701 KYM77269.1 GDQN01003078 JAT87976.1 APGK01034467 KB740914 KB631869 ENN78331.1 ERL86839.1 BT127611 AEE62573.1 JR040814 JR040815 AEY59313.1 AEY59314.1 GBYB01003039 JAG72806.1 GBYB01002739 JAG72506.1 GECU01003902 JAT03805.1 GEMB01000108 JAS03003.1 KQ983114 KYQ47311.1 GBBI01000590 JAC18122.1 GFTR01002304 JAW14122.1 KZ288262 PBC30386.1 KC507320 AHB12443.1 GDJX01024239 JAT43697.1 KQ435775 KOX75024.1 KK853170 KDR10346.1 KQ761592 OAD57369.1 GAHY01000889 JAA76621.1 EF541357 ABQ18248.1 GBHO01004326 GBRD01010559 GDHC01002003 JAG39278.1 JAG55265.1 JAQ16626.1 GBRD01010560 JAG55264.1 GDKW01003258 JAI53337.1 GEDC01011229 JAS26069.1 KJ650197 AHY30857.1 GBGD01003439 JAC85450.1 GEDC01011019 JAS26279.1 GEDC01017311 JAS19987.1 KQ434817 KZC06868.1 KQ414747 KOC61880.1 PYGN01000086 PSN54665.1 KK855850 PTY24115.1 GECU01001675 JAT06032.1 AY961556 AAX62458.1 CH902619 EDV36529.1 CVRI01000001 CRK86221.1 OUUW01000001 SPP74341.1 CH479181 EDW32366.1 CM000071 EAL25391.1 KRT02150.1 GDAI01001305 JAI16298.1 AJVK01011006 CH480824 EDW56797.1 CH954179 EDV54899.1 CM000362 CM002911 EDX08226.1 KMY95821.1 AE013599 AY113512 AAF46871.1 AAM29517.1 AY231786 AY232085 CM000158 AAR09809.1 EDW91812.1 GAMC01004690 JAC01866.1 CH916367 EDW01659.1 EZ423488 ADD19764.1 CH963849 EDW74350.1 CP012523 ALC38866.1 ATLV01024210 KE525350 KFB51000.1 GFDF01004949 JAV09135.1 GFDL01010953 JAV24092.1 DS232024 EDS32363.1
Proteomes
UP000053240
UP000007151
UP000005204
UP000053268
UP000283053
UP000037510
+ More
UP000218220 UP000005205 UP000000311 UP000036403 UP000078492 UP000007266 UP000078542 UP000008237 UP000007755 UP000078541 UP000053097 UP000078540 UP000019118 UP000030742 UP000005203 UP000075809 UP000002358 UP000242457 UP000053105 UP000027135 UP000076502 UP000053825 UP000245037 UP000192221 UP000007801 UP000183832 UP000268350 UP000008744 UP000001819 UP000092462 UP000075881 UP000001292 UP000008711 UP000000304 UP000000803 UP000002282 UP000075884 UP000001070 UP000092443 UP000091820 UP000007798 UP000092553 UP000078200 UP000092445 UP000030765 UP000002320
UP000218220 UP000005205 UP000000311 UP000036403 UP000078492 UP000007266 UP000078542 UP000008237 UP000007755 UP000078541 UP000053097 UP000078540 UP000019118 UP000030742 UP000005203 UP000075809 UP000002358 UP000242457 UP000053105 UP000027135 UP000076502 UP000053825 UP000245037 UP000192221 UP000007801 UP000183832 UP000268350 UP000008744 UP000001819 UP000092462 UP000075881 UP000001292 UP000008711 UP000000304 UP000000803 UP000002282 UP000075884 UP000001070 UP000092443 UP000091820 UP000007798 UP000092553 UP000078200 UP000092445 UP000030765 UP000002320
PRIDE
Pfam
PF01282 Ribosomal_S24e
Interpro
SUPFAM
SSF54189
SSF54189
ProteinModelPortal
I4DMD8
A0A194RK09
D1LYK8
A0A212EL84
Q6PUF7
I4DIS6
+ More
A7KCX7 G0ZED1 A0A3S2TDB1 A0A0L7K9A9 E3UKP3 A0A2W1BVS8 A0A2A4IY20 A0A0K8TH14 A0A2H1VC29 Q962Q6 S4PV71 E7DZ12 Q6F461 Q4LB02 Q4LB01 V5I9Q1 D1M7Z3 A0A158NYH2 E2A641 A0A0J7LA87 A0A151J9M0 D6WFK5 A0A195CVW2 E2C1X5 F4WT28 E9JCY5 Q4LB03 A0A195F7B6 A0A026WMG3 Q4LAZ9 A0A195AZ84 A0A1E1WM94 N6UCH0 J3JWJ3 A0A088A5I5 V9IDW0 A0A0C9QHN1 A0A0C9QGY4 A0A1B6JX74 A0A171B9G5 A0A151WHJ6 A0A023F9E9 A0A224XZS8 K7IZZ4 A0A2A3EGV2 A0A109NIA6 A0A1D1XMT2 A0A0N0U5F1 A0A067QME5 A0A310SLI5 R4FPG8 A5HMP1 A0A0A9Z7E1 A0A0K8SQ58 A0A0P4VM75 A0A1B6DK47 A0A1W5I1D7 A0A069DNQ9 A0A1B6DKS0 A0A1B6D2S6 A0A154P6P6 A0A0L7QTD8 A0A2P8ZDU5 A0A2R7WZR0 A0A1B6K3J5 Q56FE4 A0A1W4VDA6 B3MEC3 A0A1J1HE02 A0A3B0JNX3 B4GC23 Q290H1 A0A0K8TQ75 A0A1B0D3Q4 A0A182K138 B4I872 B3NLI3 B4QHJ7 Q9W229 Q6XIP3 W8BKK8 A0A182N803 B4J4H7 D3TPY6 A0A1A9XIY8 A0A1A9WNJ3 B4MQG1 A0A0M4E0V6 A0A1A9VKQ7 A0A1A9ZKW2 A0A084WLA6 A0A1L8DS36 A0A1Q3F976 B0WPH5
A7KCX7 G0ZED1 A0A3S2TDB1 A0A0L7K9A9 E3UKP3 A0A2W1BVS8 A0A2A4IY20 A0A0K8TH14 A0A2H1VC29 Q962Q6 S4PV71 E7DZ12 Q6F461 Q4LB02 Q4LB01 V5I9Q1 D1M7Z3 A0A158NYH2 E2A641 A0A0J7LA87 A0A151J9M0 D6WFK5 A0A195CVW2 E2C1X5 F4WT28 E9JCY5 Q4LB03 A0A195F7B6 A0A026WMG3 Q4LAZ9 A0A195AZ84 A0A1E1WM94 N6UCH0 J3JWJ3 A0A088A5I5 V9IDW0 A0A0C9QHN1 A0A0C9QGY4 A0A1B6JX74 A0A171B9G5 A0A151WHJ6 A0A023F9E9 A0A224XZS8 K7IZZ4 A0A2A3EGV2 A0A109NIA6 A0A1D1XMT2 A0A0N0U5F1 A0A067QME5 A0A310SLI5 R4FPG8 A5HMP1 A0A0A9Z7E1 A0A0K8SQ58 A0A0P4VM75 A0A1B6DK47 A0A1W5I1D7 A0A069DNQ9 A0A1B6DKS0 A0A1B6D2S6 A0A154P6P6 A0A0L7QTD8 A0A2P8ZDU5 A0A2R7WZR0 A0A1B6K3J5 Q56FE4 A0A1W4VDA6 B3MEC3 A0A1J1HE02 A0A3B0JNX3 B4GC23 Q290H1 A0A0K8TQ75 A0A1B0D3Q4 A0A182K138 B4I872 B3NLI3 B4QHJ7 Q9W229 Q6XIP3 W8BKK8 A0A182N803 B4J4H7 D3TPY6 A0A1A9XIY8 A0A1A9WNJ3 B4MQG1 A0A0M4E0V6 A0A1A9VKQ7 A0A1A9ZKW2 A0A084WLA6 A0A1L8DS36 A0A1Q3F976 B0WPH5
PDB
4V6W
E-value=3.21237e-46,
Score=459
Ontologies
GO
PANTHER
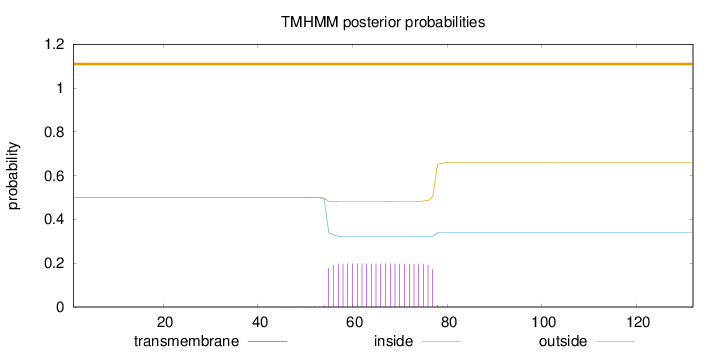
Topology
Length:
132
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
4.52068
Exp number, first 60 AAs:
1.17415
Total prob of N-in:
0.50041
outside
1 - 132
Population Genetic Test Statistics
Pi
289.922646
Theta
227.971817
Tajima's D
0.840258
CLR
0
CSRT
0.612619369031548
Interpretation
Uncertain
