Gene
KWMTBOMO10584 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003917
Annotation
neutral_lipase_[Helicoverpa_armigera]
Location in the cell
Extracellular Reliability : 2.952
Sequence
CDS
ATGAACTATTTAATTTATTGTGTTTTGTTACGTGCAGTTTGTGTTAGTGCTAATTGGGCACCATTCAAAGCGCTTTTGTATTCGGATTGGATCGAATGTAACCACAACAAGACCCTAAATTTAGGCGTAAGCGAGGTTCAAGTTTACTTCTACGACTTCCAGAACAATTTCAACACCTCATACCCGATAGATGTCGCTGTCGAGGGTATCACTTCCACCTATAACTTGGATGTGACGAGGAAACTCATCTTCTATGTACCGGGATATAAGAGTCAGATTGACAAGAACAATCCAGAACTAGTCAGACAGACGTTCAAGGACGTTCAAAACACGTATCTAATAATAATAGATCATTCCAGTTACACTTCGGACAGAGGTGGAAGATTCGAGAGCTACGAAAGATCGGTCAGTTTCGTCTATTACATTGGTAAAGCGTTGGGCAAAGTGTTGGCCGATCTACACAACAAAGGGTATCCGTCGAAAAATATACATTGTATTGGACACAGCTTGGGAGCTCAGATGTTGGGCCACGTTGGTACGACGTATACCGAATTGACCAACGAAAAAGTTTGGAGAGCCACCGGTATAGATCCGGCCGGTCCTTGCTTCTCGAATAGTTTGATTCAGGAGCAAATACGTTCCGGTGTTGCTGAATACGTTGAGGTTTACCATTGTAATGCCGGGGAACTGGGGACCACAGCCGTTCTGGCCGACATAGACTTCTTCATGAACAAGAAAGGAAAGAAACAGCCTCCATGTCACGAGGGTTTGATACCGGCGAAAGGTGATTCGGATGCCGCGAAATGCAGTCACAAAGAATGCGTGAAGTATTATATGTTGAGCGTCCACCATCCGACTTGGTATCTGGCTTGGGCGTGTGACAAATACGACGACTTCAAACATGGCAAATGCGCGGCCCATGAAGTAACAATAGCCGGGTATCACAACCCCGGCAACGCTACCGGAGTCTTCTATGTGTCAACTGAAGGATATGGGATAGATTAA
Protein
MNYLIYCVLLRAVCVSANWAPFKALLYSDWIECNHNKTLNLGVSEVQVYFYDFQNNFNTSYPIDVAVEGITSTYNLDVTRKLIFYVPGYKSQIDKNNPELVRQTFKDVQNTYLIIIDHSSYTSDRGGRFESYERSVSFVYYIGKALGKVLADLHNKGYPSKNIHCIGHSLGAQMLGHVGTTYTELTNEKVWRATGIDPAGPCFSNSLIQEQIRSGVAEYVEVYHCNAGELGTTAVLADIDFFMNKKGKKQPPCHEGLIPAKGDSDAAKCSHKECVKYYMLSVHHPTWYLAWACDKYDDFKHGKCAAHEVTIAGYHNPGNATGVFYVSTEGYGID
Summary
Similarity
Belongs to the AB hydrolase superfamily. Lipase family.
Uniprot
H9J330
I3QC15
A0A2H1V1J5
A0A0L7KP67
A0A194PIB5
A0A194PIK8
+ More
A0A3S2KZ92 A0A194RJD6 A0A212EL87 A0A194RJQ0 A0A212EL72 A0A2A4J4J5 A0A194PJD2 A0A1E1WHL6 A0A2A4J3L7 A0A2W1BRK5 A0A2H1V5T0 H9J333 A0A1E1WCM9 A0A2W1BTP2 A0A194QHM4 A0A0L7KP87 A0A2H1W5A8 A0A2W1C0X1 A0A2H1V5Q6 A0A2W1BWG2 A0A2W1BXY3 A0A3S2NZZ7 A0A2H1WNR8 A0A0L7L8R4 A0A220K8L9 A0A2A4J5Z0 B4IEY9 A0A3S2L3L9 A0A212EJL3 A0A2W1BV63 B3NWD0 Q06VE9 A0A2N9QV27 B3MW48 B4Q270 A0A2H1W7F1 A0A2H1W7Q1 A0A2A4IT83 A0A1I8Q014 B4R6E3 Q9VX70 A0A212FKA5 I6TFN4 A0A087ZXA0 H9JWY8 A0A0L0CRF9 A0A194QC74 A0A2A4JGD9 A0A2H1X3Q9 A0A194RQL7 B4NC73 A0A146LFH2 A0A1I8MA26 A0A194PSE8 A0A034WKC4 A0A0L7KP71 A0A1W4VWC2 A0A0K8VXK1 A0A0N0PB11 B4L3H4 Q9VPH3 A0A2H1WL84 A0A0A9XTP0 A0A0A1WYA6 A0A2S2QF57 A0A0L0C2D6 A0A034VT37 A0A0K8U3C5 E9IDK7 B3M978 U5EEG1 E2AIM4 B4QRX5 B5DM35 B3NIK0 B4IA88 A0A1I8N5B5 A0A0K8UZ74 A0A191XQW7 B4GY97 A0A2W1B7Q2 B4PFA3
A0A3S2KZ92 A0A194RJD6 A0A212EL87 A0A194RJQ0 A0A212EL72 A0A2A4J4J5 A0A194PJD2 A0A1E1WHL6 A0A2A4J3L7 A0A2W1BRK5 A0A2H1V5T0 H9J333 A0A1E1WCM9 A0A2W1BTP2 A0A194QHM4 A0A0L7KP87 A0A2H1W5A8 A0A2W1C0X1 A0A2H1V5Q6 A0A2W1BWG2 A0A2W1BXY3 A0A3S2NZZ7 A0A2H1WNR8 A0A0L7L8R4 A0A220K8L9 A0A2A4J5Z0 B4IEY9 A0A3S2L3L9 A0A212EJL3 A0A2W1BV63 B3NWD0 Q06VE9 A0A2N9QV27 B3MW48 B4Q270 A0A2H1W7F1 A0A2H1W7Q1 A0A2A4IT83 A0A1I8Q014 B4R6E3 Q9VX70 A0A212FKA5 I6TFN4 A0A087ZXA0 H9JWY8 A0A0L0CRF9 A0A194QC74 A0A2A4JGD9 A0A2H1X3Q9 A0A194RQL7 B4NC73 A0A146LFH2 A0A1I8MA26 A0A194PSE8 A0A034WKC4 A0A0L7KP71 A0A1W4VWC2 A0A0K8VXK1 A0A0N0PB11 B4L3H4 Q9VPH3 A0A2H1WL84 A0A0A9XTP0 A0A0A1WYA6 A0A2S2QF57 A0A0L0C2D6 A0A034VT37 A0A0K8U3C5 E9IDK7 B3M978 U5EEG1 E2AIM4 B4QRX5 B5DM35 B3NIK0 B4IA88 A0A1I8N5B5 A0A0K8UZ74 A0A191XQW7 B4GY97 A0A2W1B7Q2 B4PFA3
Pubmed
EMBL
BABH01037645
JF999964
KZ149921
AFI64318.1
PZC77724.1
ODYU01000260
+ More
SOQ34725.1 JTDY01007577 KOB65103.1 KQ459603 KPI93151.1 KPI93152.1 RSAL01000385 RVE42038.1 KQ460124 KPJ17667.1 AGBW02014130 OWR42231.1 KPJ17664.1 OWR42232.1 NWSH01003264 PCG66656.1 KPI93148.1 GDQN01004565 JAT86489.1 PCG66655.1 PZC77722.1 ODYU01000811 SOQ36159.1 GDQN01006349 JAT84705.1 PZC77721.1 KQ459220 KPJ02946.1 JTDY01007571 KOB65107.1 ODYU01006304 SOQ48032.1 PZC77723.1 SOQ36158.1 KZ149926 PZC77547.1 PZC77546.1 RSAL01000012 RVE53468.1 ODYU01009953 SOQ54715.1 JTDY01002277 KOB71750.1 MF319718 ASJ26432.1 NWSH01003103 PCG66944.1 CH480832 EDW46243.1 RSAL01000184 RVE44850.1 AGBW02014435 OWR41693.1 PZC77545.1 CH954180 EDV46469.1 DQ517337 ABF70649.1 KY434117 AUS94238.1 CH902625 EDV35193.1 CM000162 EDX02578.1 ODYU01006796 SOQ48977.1 SOQ48976.1 NWSH01007523 PCG62931.1 CM000366 EDX18193.1 AE014298 BT024983 AAF48708.2 ABE01213.1 AGBW02008082 OWR54177.1 JQ814807 AFM73623.1 BABH01041781 JRES01000120 KNC34019.1 KQ459193 KPJ03024.1 NWSH01001670 PCG70462.1 ODYU01013233 SOQ59929.1 KQ459833 KPJ19782.1 CH964239 EDW82432.2 GDHC01012863 JAQ05766.1 KQ459601 KPI94050.1 GAKP01004764 JAC54188.1 JTDY01007594 KOB65092.1 GDHF01008697 JAI43617.1 KQ461072 KPJ09835.1 CH933810 EDW07102.1 AE014296 AAF51579.2 ODYU01009427 SOQ53845.1 GBHO01020355 JAG23249.1 GBXI01010636 JAD03656.1 GGMS01007165 MBY76368.1 JRES01000987 KNC26392.1 GAKP01013338 JAC45614.1 GDHF01031494 JAI20820.1 GL762454 EFZ21361.1 CH902618 EDV40062.2 GANO01004326 JAB55545.1 GL439840 EFN66713.1 CM000363 CM002912 EDX11221.1 KMZ00758.1 CH379064 EDY72552.1 CH954178 EDV52496.1 CH480826 EDW44201.1 GDHF01020674 JAI31640.1 KU360126 ANJ42864.1 CH479197 EDW27753.1 KZ150304 PZC71518.1 CM000159 EDW95189.1
SOQ34725.1 JTDY01007577 KOB65103.1 KQ459603 KPI93151.1 KPI93152.1 RSAL01000385 RVE42038.1 KQ460124 KPJ17667.1 AGBW02014130 OWR42231.1 KPJ17664.1 OWR42232.1 NWSH01003264 PCG66656.1 KPI93148.1 GDQN01004565 JAT86489.1 PCG66655.1 PZC77722.1 ODYU01000811 SOQ36159.1 GDQN01006349 JAT84705.1 PZC77721.1 KQ459220 KPJ02946.1 JTDY01007571 KOB65107.1 ODYU01006304 SOQ48032.1 PZC77723.1 SOQ36158.1 KZ149926 PZC77547.1 PZC77546.1 RSAL01000012 RVE53468.1 ODYU01009953 SOQ54715.1 JTDY01002277 KOB71750.1 MF319718 ASJ26432.1 NWSH01003103 PCG66944.1 CH480832 EDW46243.1 RSAL01000184 RVE44850.1 AGBW02014435 OWR41693.1 PZC77545.1 CH954180 EDV46469.1 DQ517337 ABF70649.1 KY434117 AUS94238.1 CH902625 EDV35193.1 CM000162 EDX02578.1 ODYU01006796 SOQ48977.1 SOQ48976.1 NWSH01007523 PCG62931.1 CM000366 EDX18193.1 AE014298 BT024983 AAF48708.2 ABE01213.1 AGBW02008082 OWR54177.1 JQ814807 AFM73623.1 BABH01041781 JRES01000120 KNC34019.1 KQ459193 KPJ03024.1 NWSH01001670 PCG70462.1 ODYU01013233 SOQ59929.1 KQ459833 KPJ19782.1 CH964239 EDW82432.2 GDHC01012863 JAQ05766.1 KQ459601 KPI94050.1 GAKP01004764 JAC54188.1 JTDY01007594 KOB65092.1 GDHF01008697 JAI43617.1 KQ461072 KPJ09835.1 CH933810 EDW07102.1 AE014296 AAF51579.2 ODYU01009427 SOQ53845.1 GBHO01020355 JAG23249.1 GBXI01010636 JAD03656.1 GGMS01007165 MBY76368.1 JRES01000987 KNC26392.1 GAKP01013338 JAC45614.1 GDHF01031494 JAI20820.1 GL762454 EFZ21361.1 CH902618 EDV40062.2 GANO01004326 JAB55545.1 GL439840 EFN66713.1 CM000363 CM002912 EDX11221.1 KMZ00758.1 CH379064 EDY72552.1 CH954178 EDV52496.1 CH480826 EDW44201.1 GDHF01020674 JAI31640.1 KU360126 ANJ42864.1 CH479197 EDW27753.1 KZ150304 PZC71518.1 CM000159 EDW95189.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
H9J330
I3QC15
A0A2H1V1J5
A0A0L7KP67
A0A194PIB5
A0A194PIK8
+ More
A0A3S2KZ92 A0A194RJD6 A0A212EL87 A0A194RJQ0 A0A212EL72 A0A2A4J4J5 A0A194PJD2 A0A1E1WHL6 A0A2A4J3L7 A0A2W1BRK5 A0A2H1V5T0 H9J333 A0A1E1WCM9 A0A2W1BTP2 A0A194QHM4 A0A0L7KP87 A0A2H1W5A8 A0A2W1C0X1 A0A2H1V5Q6 A0A2W1BWG2 A0A2W1BXY3 A0A3S2NZZ7 A0A2H1WNR8 A0A0L7L8R4 A0A220K8L9 A0A2A4J5Z0 B4IEY9 A0A3S2L3L9 A0A212EJL3 A0A2W1BV63 B3NWD0 Q06VE9 A0A2N9QV27 B3MW48 B4Q270 A0A2H1W7F1 A0A2H1W7Q1 A0A2A4IT83 A0A1I8Q014 B4R6E3 Q9VX70 A0A212FKA5 I6TFN4 A0A087ZXA0 H9JWY8 A0A0L0CRF9 A0A194QC74 A0A2A4JGD9 A0A2H1X3Q9 A0A194RQL7 B4NC73 A0A146LFH2 A0A1I8MA26 A0A194PSE8 A0A034WKC4 A0A0L7KP71 A0A1W4VWC2 A0A0K8VXK1 A0A0N0PB11 B4L3H4 Q9VPH3 A0A2H1WL84 A0A0A9XTP0 A0A0A1WYA6 A0A2S2QF57 A0A0L0C2D6 A0A034VT37 A0A0K8U3C5 E9IDK7 B3M978 U5EEG1 E2AIM4 B4QRX5 B5DM35 B3NIK0 B4IA88 A0A1I8N5B5 A0A0K8UZ74 A0A191XQW7 B4GY97 A0A2W1B7Q2 B4PFA3
A0A3S2KZ92 A0A194RJD6 A0A212EL87 A0A194RJQ0 A0A212EL72 A0A2A4J4J5 A0A194PJD2 A0A1E1WHL6 A0A2A4J3L7 A0A2W1BRK5 A0A2H1V5T0 H9J333 A0A1E1WCM9 A0A2W1BTP2 A0A194QHM4 A0A0L7KP87 A0A2H1W5A8 A0A2W1C0X1 A0A2H1V5Q6 A0A2W1BWG2 A0A2W1BXY3 A0A3S2NZZ7 A0A2H1WNR8 A0A0L7L8R4 A0A220K8L9 A0A2A4J5Z0 B4IEY9 A0A3S2L3L9 A0A212EJL3 A0A2W1BV63 B3NWD0 Q06VE9 A0A2N9QV27 B3MW48 B4Q270 A0A2H1W7F1 A0A2H1W7Q1 A0A2A4IT83 A0A1I8Q014 B4R6E3 Q9VX70 A0A212FKA5 I6TFN4 A0A087ZXA0 H9JWY8 A0A0L0CRF9 A0A194QC74 A0A2A4JGD9 A0A2H1X3Q9 A0A194RQL7 B4NC73 A0A146LFH2 A0A1I8MA26 A0A194PSE8 A0A034WKC4 A0A0L7KP71 A0A1W4VWC2 A0A0K8VXK1 A0A0N0PB11 B4L3H4 Q9VPH3 A0A2H1WL84 A0A0A9XTP0 A0A0A1WYA6 A0A2S2QF57 A0A0L0C2D6 A0A034VT37 A0A0K8U3C5 E9IDK7 B3M978 U5EEG1 E2AIM4 B4QRX5 B5DM35 B3NIK0 B4IA88 A0A1I8N5B5 A0A0K8UZ74 A0A191XQW7 B4GY97 A0A2W1B7Q2 B4PFA3
PDB
1W52
E-value=3.86165e-23,
Score=267
Ontologies
GO
PANTHER
Topology
Subcellular location
Secreted
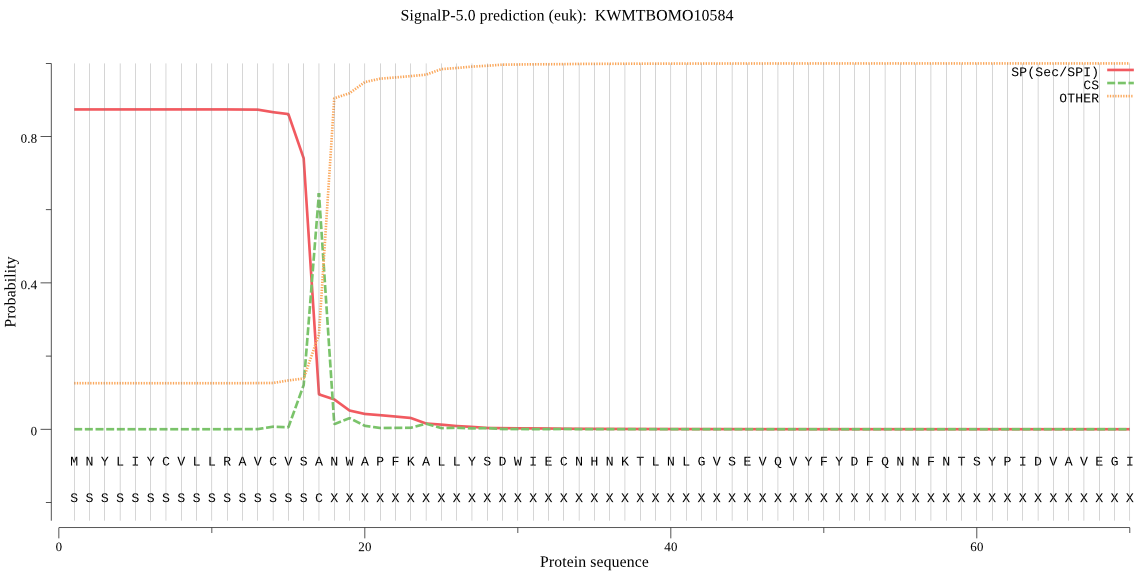
SignalP
Position: 1 - 17,
Likelihood: 0.873376
Length:
334
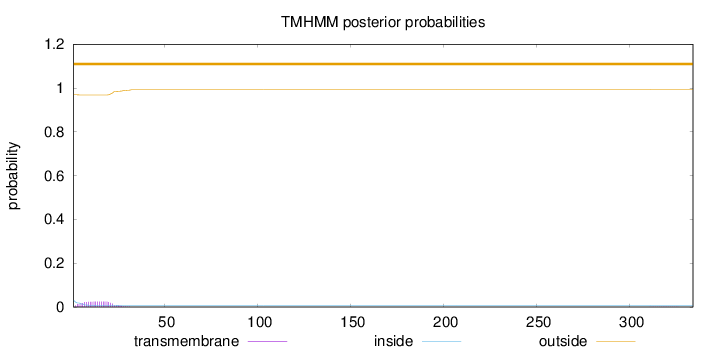
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.490789999999999
Exp number, first 60 AAs:
0.48469
Total prob of N-in:
0.03119
outside
1 - 334
Population Genetic Test Statistics
Pi
196.951629
Theta
148.224815
Tajima's D
1.367684
CLR
0.023221
CSRT
0.757462126893655
Interpretation
Uncertain
