Gene
KWMTBOMO10583
Pre Gene Modal
BGIBMGA003920
Annotation
PREDICTED:_pancreatic_lipase-related_protein_2-like_[Papilio_machaon]
Location in the cell
Extracellular Reliability : 1.021 Lysosomal Reliability : 1.199 Nuclear Reliability : 1.024
Sequence
CDS
ATGTTGGATCATTCTCTATACACCAACAACAAACAAGGCAATAGGAAGAGCTACGAGAGATCTGTTAAGTACGTTCACTATATTGGTAAAGCGTTGGGACAAATGTTGGTTAAAATTCGTGACGGCGGTGTAACCACACAGAAAATGCACTGTATTGGCCACAGTCTTGGAGCGCAAATATTGGGACATGCTGGCGATGAATTCATGAGGCTGGGCTTTGAGAGAATACAGAGGATCACCGCTTTAGACCCAGCCGGACCTTGCTTTTCGAACAGTTTTATTGAGGAGCAGGTTAGAGCTGGTGTGGCGAAATACGTTGAGGTCTACCACTGCAATGCTGGCGGTTTGGGTAGTTCCAGCGTATTAGGCGATGTGGACTTCTTCATTAACAAAAACGGACAAACCCAACCCAACTGCAGCACTCCTTGGATTCCCGGCATCTTTGACTCGCCAAAAGCGGCTTCGTGCAATCACAGAACTTGCATTGACATTTGGACTGCGTCAGTGAGACATCCAGAATGGTTCCCGGCATGGAAATGCAACTCGTATAAGGACTTCAAGAAGGGAAGATGTGCAACCAGCGAAATGTCCATAGCCGGCTATTGGAATCCTGGAAATGCAACTGGTATATACTACTTCAAGACCGATGGCTACAATCTGAGTTCATAA
Protein
MLDHSLYTNNKQGNRKSYERSVKYVHYIGKALGQMLVKIRDGGVTTQKMHCIGHSLGAQILGHAGDEFMRLGFERIQRITALDPAGPCFSNSFIEEQVRAGVAKYVEVYHCNAGGLGSSSVLGDVDFFINKNGQTQPNCSTPWIPGIFDSPKAASCNHRTCIDIWTASVRHPEWFPAWKCNSYKDFKKGRCATSEMSIAGYWNPGNATGIYYFKTDGYNLSS
Summary
Similarity
Belongs to the AB hydrolase superfamily. Lipase family.
Uniprot
H9J333
A0A194RJQ0
A0A194PJD2
A0A212EL72
A0A1E1WCM9
A0A2H1V5T0
+ More
A0A2W1BRK5 A0A2A4J3L7 A0A2A4J4J5 A0A3S2KZ92 A0A0L7KP87 A0A0L7KP67 A0A194PIB5 A0A194PIK8 A0A2W1BTP2 I3QC15 A0A194RJD6 A0A1E1WHL6 A0A212EL87 A0A2H1V1J5 H9J330 A0A2H1W5A8 A0A2H1V5Q6 A0A2W1C0X1 A0A0N1IQQ4 A0A194QHM4 A0A2W1BWG2 A0A220K8L9 A0A2A4J690 A0A2A4JYV7 A0A3S2NZZ7 A0A212EJL3 A0A0L7L8R4 A0A2H1W7Q1 A0A2W1BXY3 A0A2A4JGD9 A0A3S2L3L9 A0A0L7KP71 I6TFN4 H9JWY8 A0A2H1W7F1 A0A2H1X3Q9 A0A194PSE8 A0A194RQL7 A0A194QXT2 A0A2A4J5Z0 A0A2H1W795 A0A2W1BV63 A0A212FKA5 H9JFT5 A0A2H1WL84 A0A194QNS4 A0A2H1WNR8 K7IR20 A0A2H1VJ13 A0A1Y1NEG9 A0A194PCA8 F4WY21 A0A158NYL0 B4IEY9 A0A232F8C9 A0A191XQW7 A0A2A4IT83 A0A194QC74 A0A0K8SS21 H9JLK0 A0A2H1VGU5 A0A1J1IUY9 A0A0A9X8M8 A0A146LJU7 A0A0K8SRI3 A0A0N1II42 A0A195FVA2 A0A0N0PB11 B4R6E3 D6WU98 Q9VX70 A0A194Q1D6 A0A0P4VMV4 A0A2N9QV27 A0A2A4JBH9 Q06VE9 G3VIQ6 A0A1J1ICM9 K1QQ27 A0A0K8WHX4 A0A1I8Q014 A0A067QXJ2 A0A1J1J6D0 U5EEG1 A0A087ZXA4 A0A3S2LBT4 E2A4Y7 A0A023ES65 A0A182GQ76 S4RQL2
A0A2W1BRK5 A0A2A4J3L7 A0A2A4J4J5 A0A3S2KZ92 A0A0L7KP87 A0A0L7KP67 A0A194PIB5 A0A194PIK8 A0A2W1BTP2 I3QC15 A0A194RJD6 A0A1E1WHL6 A0A212EL87 A0A2H1V1J5 H9J330 A0A2H1W5A8 A0A2H1V5Q6 A0A2W1C0X1 A0A0N1IQQ4 A0A194QHM4 A0A2W1BWG2 A0A220K8L9 A0A2A4J690 A0A2A4JYV7 A0A3S2NZZ7 A0A212EJL3 A0A0L7L8R4 A0A2H1W7Q1 A0A2W1BXY3 A0A2A4JGD9 A0A3S2L3L9 A0A0L7KP71 I6TFN4 H9JWY8 A0A2H1W7F1 A0A2H1X3Q9 A0A194PSE8 A0A194RQL7 A0A194QXT2 A0A2A4J5Z0 A0A2H1W795 A0A2W1BV63 A0A212FKA5 H9JFT5 A0A2H1WL84 A0A194QNS4 A0A2H1WNR8 K7IR20 A0A2H1VJ13 A0A1Y1NEG9 A0A194PCA8 F4WY21 A0A158NYL0 B4IEY9 A0A232F8C9 A0A191XQW7 A0A2A4IT83 A0A194QC74 A0A0K8SS21 H9JLK0 A0A2H1VGU5 A0A1J1IUY9 A0A0A9X8M8 A0A146LJU7 A0A0K8SRI3 A0A0N1II42 A0A195FVA2 A0A0N0PB11 B4R6E3 D6WU98 Q9VX70 A0A194Q1D6 A0A0P4VMV4 A0A2N9QV27 A0A2A4JBH9 Q06VE9 G3VIQ6 A0A1J1ICM9 K1QQ27 A0A0K8WHX4 A0A1I8Q014 A0A067QXJ2 A0A1J1J6D0 U5EEG1 A0A087ZXA4 A0A3S2LBT4 E2A4Y7 A0A023ES65 A0A182GQ76 S4RQL2
Pubmed
19121390
26354079
22118469
28756777
26227816
28630352
+ More
23171126 20075255 28004739 21719571 21347285 17994087 28648823 25401762 26823975 18362917 19820115 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 27129103 16876847 21709235 22992520 24845553 20798317 24945155 26483478
23171126 20075255 28004739 21719571 21347285 17994087 28648823 25401762 26823975 18362917 19820115 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 27129103 16876847 21709235 22992520 24845553 20798317 24945155 26483478
EMBL
BABH01037645
KQ460124
KPJ17664.1
KQ459603
KPI93148.1
AGBW02014130
+ More
OWR42232.1 GDQN01006349 JAT84705.1 ODYU01000811 SOQ36159.1 KZ149921 PZC77722.1 NWSH01003264 PCG66655.1 PCG66656.1 RSAL01000385 RVE42038.1 JTDY01007571 KOB65107.1 JTDY01007577 KOB65103.1 KPI93151.1 KPI93152.1 PZC77721.1 JF999964 AFI64318.1 PZC77724.1 KPJ17667.1 GDQN01004565 JAT86489.1 OWR42231.1 ODYU01000260 SOQ34725.1 ODYU01006304 SOQ48032.1 SOQ36158.1 PZC77723.1 LADJ01052308 KPJ21527.1 KQ459220 KPJ02946.1 KZ149926 PZC77547.1 MF319718 ASJ26432.1 NWSH01003103 PCG66943.1 NWSH01000413 PCG76562.1 RSAL01000012 RVE53468.1 AGBW02014435 OWR41693.1 JTDY01002277 KOB71750.1 ODYU01006796 SOQ48976.1 PZC77546.1 NWSH01001670 PCG70462.1 RSAL01000184 RVE44850.1 JTDY01007594 KOB65092.1 JQ814807 AFM73623.1 BABH01041781 SOQ48977.1 ODYU01013233 SOQ59929.1 KQ459601 KPI94050.1 KQ459833 KPJ19782.1 KQ461154 KPJ08386.1 PCG66944.1 SOQ48975.1 PZC77545.1 AGBW02008082 OWR54177.1 BABH01001653 ODYU01009427 SOQ53845.1 KQ461196 KPJ06625.1 ODYU01009953 SOQ54715.1 ODYU01002823 SOQ40786.1 GEZM01005476 JAV96149.1 KQ459606 KPI90911.1 GL888434 EGI60946.1 ADTU01004020 CH480832 EDW46243.1 NNAY01000669 OXU27086.1 KU360126 ANJ42864.1 NWSH01007523 PCG62931.1 KQ459193 KPJ03024.1 GBRD01009843 JAG55981.1 BABH01005602 ODYU01002500 SOQ40069.1 CVRI01000061 CRL03999.1 GBHO01027613 JAG15991.1 GDHC01011552 JAQ07077.1 GBRD01009842 JAG55982.1 KQ460391 KPJ15413.1 KQ981276 KYN43794.1 KQ461072 KPJ09835.1 CM000366 EDX18193.1 KQ971352 EFA06788.1 AE014298 BT024983 AAF48708.2 ABE01213.1 KQ459590 KPI97210.1 GDKW01002938 JAI53657.1 KY434117 AUS94238.1 NWSH01002209 PCG68904.1 DQ517337 ABF70649.1 AEFK01140443 AEFK01140444 CVRI01000047 CRK97955.1 JH816729 EKC35943.1 GDHF01001578 JAI50736.1 KK852855 KDR14939.1 CVRI01000074 CRL07959.1 GANO01004326 JAB55545.1 RSAL01000052 RVE50204.1 GL436770 EFN71504.1 GAPW01002017 JAC11581.1 JXUM01080013 JXUM01080014 KQ563159 KXJ74375.1
OWR42232.1 GDQN01006349 JAT84705.1 ODYU01000811 SOQ36159.1 KZ149921 PZC77722.1 NWSH01003264 PCG66655.1 PCG66656.1 RSAL01000385 RVE42038.1 JTDY01007571 KOB65107.1 JTDY01007577 KOB65103.1 KPI93151.1 KPI93152.1 PZC77721.1 JF999964 AFI64318.1 PZC77724.1 KPJ17667.1 GDQN01004565 JAT86489.1 OWR42231.1 ODYU01000260 SOQ34725.1 ODYU01006304 SOQ48032.1 SOQ36158.1 PZC77723.1 LADJ01052308 KPJ21527.1 KQ459220 KPJ02946.1 KZ149926 PZC77547.1 MF319718 ASJ26432.1 NWSH01003103 PCG66943.1 NWSH01000413 PCG76562.1 RSAL01000012 RVE53468.1 AGBW02014435 OWR41693.1 JTDY01002277 KOB71750.1 ODYU01006796 SOQ48976.1 PZC77546.1 NWSH01001670 PCG70462.1 RSAL01000184 RVE44850.1 JTDY01007594 KOB65092.1 JQ814807 AFM73623.1 BABH01041781 SOQ48977.1 ODYU01013233 SOQ59929.1 KQ459601 KPI94050.1 KQ459833 KPJ19782.1 KQ461154 KPJ08386.1 PCG66944.1 SOQ48975.1 PZC77545.1 AGBW02008082 OWR54177.1 BABH01001653 ODYU01009427 SOQ53845.1 KQ461196 KPJ06625.1 ODYU01009953 SOQ54715.1 ODYU01002823 SOQ40786.1 GEZM01005476 JAV96149.1 KQ459606 KPI90911.1 GL888434 EGI60946.1 ADTU01004020 CH480832 EDW46243.1 NNAY01000669 OXU27086.1 KU360126 ANJ42864.1 NWSH01007523 PCG62931.1 KQ459193 KPJ03024.1 GBRD01009843 JAG55981.1 BABH01005602 ODYU01002500 SOQ40069.1 CVRI01000061 CRL03999.1 GBHO01027613 JAG15991.1 GDHC01011552 JAQ07077.1 GBRD01009842 JAG55982.1 KQ460391 KPJ15413.1 KQ981276 KYN43794.1 KQ461072 KPJ09835.1 CM000366 EDX18193.1 KQ971352 EFA06788.1 AE014298 BT024983 AAF48708.2 ABE01213.1 KQ459590 KPI97210.1 GDKW01002938 JAI53657.1 KY434117 AUS94238.1 NWSH01002209 PCG68904.1 DQ517337 ABF70649.1 AEFK01140443 AEFK01140444 CVRI01000047 CRK97955.1 JH816729 EKC35943.1 GDHF01001578 JAI50736.1 KK852855 KDR14939.1 CVRI01000074 CRL07959.1 GANO01004326 JAB55545.1 RSAL01000052 RVE50204.1 GL436770 EFN71504.1 GAPW01002017 JAC11581.1 JXUM01080013 JXUM01080014 KQ563159 KXJ74375.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000218220
UP000283053
+ More
UP000037510 UP000002358 UP000007755 UP000005205 UP000001292 UP000215335 UP000183832 UP000078541 UP000000304 UP000007266 UP000000803 UP000001323 UP000007648 UP000005408 UP000095300 UP000027135 UP000005203 UP000000311 UP000069940 UP000249989 UP000245300
UP000037510 UP000002358 UP000007755 UP000005205 UP000001292 UP000215335 UP000183832 UP000078541 UP000000304 UP000007266 UP000000803 UP000001323 UP000007648 UP000005408 UP000095300 UP000027135 UP000005203 UP000000311 UP000069940 UP000249989 UP000245300
Interpro
Gene 3D
ProteinModelPortal
H9J333
A0A194RJQ0
A0A194PJD2
A0A212EL72
A0A1E1WCM9
A0A2H1V5T0
+ More
A0A2W1BRK5 A0A2A4J3L7 A0A2A4J4J5 A0A3S2KZ92 A0A0L7KP87 A0A0L7KP67 A0A194PIB5 A0A194PIK8 A0A2W1BTP2 I3QC15 A0A194RJD6 A0A1E1WHL6 A0A212EL87 A0A2H1V1J5 H9J330 A0A2H1W5A8 A0A2H1V5Q6 A0A2W1C0X1 A0A0N1IQQ4 A0A194QHM4 A0A2W1BWG2 A0A220K8L9 A0A2A4J690 A0A2A4JYV7 A0A3S2NZZ7 A0A212EJL3 A0A0L7L8R4 A0A2H1W7Q1 A0A2W1BXY3 A0A2A4JGD9 A0A3S2L3L9 A0A0L7KP71 I6TFN4 H9JWY8 A0A2H1W7F1 A0A2H1X3Q9 A0A194PSE8 A0A194RQL7 A0A194QXT2 A0A2A4J5Z0 A0A2H1W795 A0A2W1BV63 A0A212FKA5 H9JFT5 A0A2H1WL84 A0A194QNS4 A0A2H1WNR8 K7IR20 A0A2H1VJ13 A0A1Y1NEG9 A0A194PCA8 F4WY21 A0A158NYL0 B4IEY9 A0A232F8C9 A0A191XQW7 A0A2A4IT83 A0A194QC74 A0A0K8SS21 H9JLK0 A0A2H1VGU5 A0A1J1IUY9 A0A0A9X8M8 A0A146LJU7 A0A0K8SRI3 A0A0N1II42 A0A195FVA2 A0A0N0PB11 B4R6E3 D6WU98 Q9VX70 A0A194Q1D6 A0A0P4VMV4 A0A2N9QV27 A0A2A4JBH9 Q06VE9 G3VIQ6 A0A1J1ICM9 K1QQ27 A0A0K8WHX4 A0A1I8Q014 A0A067QXJ2 A0A1J1J6D0 U5EEG1 A0A087ZXA4 A0A3S2LBT4 E2A4Y7 A0A023ES65 A0A182GQ76 S4RQL2
A0A2W1BRK5 A0A2A4J3L7 A0A2A4J4J5 A0A3S2KZ92 A0A0L7KP87 A0A0L7KP67 A0A194PIB5 A0A194PIK8 A0A2W1BTP2 I3QC15 A0A194RJD6 A0A1E1WHL6 A0A212EL87 A0A2H1V1J5 H9J330 A0A2H1W5A8 A0A2H1V5Q6 A0A2W1C0X1 A0A0N1IQQ4 A0A194QHM4 A0A2W1BWG2 A0A220K8L9 A0A2A4J690 A0A2A4JYV7 A0A3S2NZZ7 A0A212EJL3 A0A0L7L8R4 A0A2H1W7Q1 A0A2W1BXY3 A0A2A4JGD9 A0A3S2L3L9 A0A0L7KP71 I6TFN4 H9JWY8 A0A2H1W7F1 A0A2H1X3Q9 A0A194PSE8 A0A194RQL7 A0A194QXT2 A0A2A4J5Z0 A0A2H1W795 A0A2W1BV63 A0A212FKA5 H9JFT5 A0A2H1WL84 A0A194QNS4 A0A2H1WNR8 K7IR20 A0A2H1VJ13 A0A1Y1NEG9 A0A194PCA8 F4WY21 A0A158NYL0 B4IEY9 A0A232F8C9 A0A191XQW7 A0A2A4IT83 A0A194QC74 A0A0K8SS21 H9JLK0 A0A2H1VGU5 A0A1J1IUY9 A0A0A9X8M8 A0A146LJU7 A0A0K8SRI3 A0A0N1II42 A0A195FVA2 A0A0N0PB11 B4R6E3 D6WU98 Q9VX70 A0A194Q1D6 A0A0P4VMV4 A0A2N9QV27 A0A2A4JBH9 Q06VE9 G3VIQ6 A0A1J1ICM9 K1QQ27 A0A0K8WHX4 A0A1I8Q014 A0A067QXJ2 A0A1J1J6D0 U5EEG1 A0A087ZXA4 A0A3S2LBT4 E2A4Y7 A0A023ES65 A0A182GQ76 S4RQL2
PDB
2PPL
E-value=1.0693e-14,
Score=192
Ontologies
GO
PANTHER
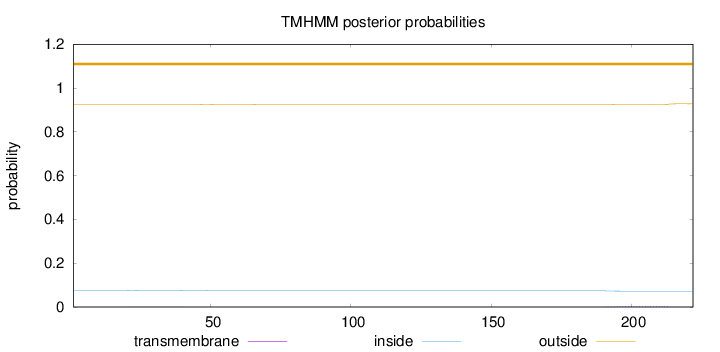
Topology
Subcellular location
Secreted
Length:
222
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.08967
Exp number, first 60 AAs:
0.01368
Total prob of N-in:
0.07537
outside
1 - 222
Population Genetic Test Statistics
Pi
167.19861
Theta
148.375113
Tajima's D
0.465752
CLR
0
CSRT
0.508374581270936
Interpretation
Uncertain
