Gene
KWMTBOMO10562 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA014031
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_15-hydroxyprostaglandin_dehydrogenase_[NAD(+)]-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.57
Sequence
CDS
ATGGCTGTAGAGTTAACGTTTCTATTTTGCCTAGTAATACTTTCGGTTAGGGCCAATTCAATCAGTGACAAGGATCTAAAAGGTAAAAATGTTATCGTGACCGGGGCAGCTAGAGGTATAGGATACGCTATAGCTGATAATTTCTTAGCTAATGGAGTCAATCTAATTATAATCTTAGATAAAAATGTGACTAACGGAGTAAGAGCAGCTGAAACTCTAAAATGTAAATATGGGAGAGGTAAGACTATTTTTATACCATGCGATGTTACGAAAGATCTTCAACTGTGGGATGGAATAATTAAAAAACATGGGCCGATACACGTGTTAGTCAATAATGCTGGAATACTTGATGAAGACCAACCAAGAGAAATGATATTGACCAACAGCGTTGCACCGATTGAATGGTCACTAAAATTCAGAGAATACGCGAGAAACGACAAAGGTGGTCCTGGTGGAACTATTATAAATACCGCGTCCAGGTCAGGTTACAGAATAACTCCGTTCATGGTTTCCTACGTCTCATCAAAGCACGCGAGTCTAGCTTTCTCCAAGTCATTAGGACACCCATACAACTTCAAAAGAACAGGGATTAGGATTGTGGCTCTATGTCCAGAACTGACTTACACTGCGATGACTGCAAAGCAAACGGTTTGGGATGATCAGTTAGAAGATTATAAAAAAGTCGTCAAAGATGCCGTTTGGCAGAAACCCGATGTTGTCGGCAAAGCAGCTGTGCACATATTCAAGAATGCCGAATCCGGGACCGCTTGGAGAATCGCTGGTGGTCACCTGAGTGTAGCTCCAGTCTATTTATACATGACTGATGAAGAAATTGAAAAAGCTATAAAACAAAATTAA
Protein
MAVELTFLFCLVILSVRANSISDKDLKGKNVIVTGAARGIGYAIADNFLANGVNLIIILDKNVTNGVRAAETLKCKYGRGKTIFIPCDVTKDLQLWDGIIKKHGPIHVLVNNAGILDEDQPREMILTNSVAPIEWSLKFREYARNDKGGPGGTIINTASRSGYRITPFMVSYVSSKHASLAFSKSLGHPYNFKRTGIRIVALCPELTYTAMTAKQTVWDDQLEDYKKVVKDAVWQKPDVVGKAAVHIFKNAESGTAWRIAGGHLSVAPVYLYMTDEEIEKAIKQN
Summary
Similarity
Belongs to the short-chain dehydrogenases/reductases (SDR) family.
Uniprot
H9JWW7
H9JWW0
H9JYK0
H9JWW1
H9JWW6
H9J8J0
+ More
Q1HPX6 A0A194R949 H9J8I9 A0A194R584 H9J369 A0A0L7KQW0 A0A0H4TDA9 A0A194R4F0 H9J8I4 A0A2H1VNC8 A0A0F6Q2J9 A0A0L7LJP6 A0A2W1BR60 A0A0F6Q0W0 H9J8I5 A0A1L8D6N5 A0A212F936 A0A194QJQ6 A0A2W1BL60 A0A2A4JIR0 A0A194R3L5 A0A2W1BSK9 A0A0F6Q0X5 S4PIB4 H9J355 U5KC20 I4DKW3 A0A0H4TH26 D2SNK3 A0A212F911 H9J8I6 A0A0H4TD16 A0A0F6Q4H7 A0A2H1VSR5 A0A0L7K2F9 H9J354 A0A2H1VR53 A0A2A4JJW8 H9J353 H9J0A6 A0A2H8TFE7 A0A2S2NV02 J9JKN0 F4W474 A0A0C9R6P4 A0A026WTY5 A0A151WLU8 K7IV86 A0A232EGH1 A0A2P8Z2R9 A0A1W4WBZ1 A0A195ET92 A0A026WRA5 A0A0J7L0X1 Q6RUR3 A0A067QWG1 A0A151X1L8 E2B6N3 A0A0L7QSX6 A0A0J7L5T5 A0A140KPR8 B0XI43 A0A195B3G6 A0A195B441 A0A212F0F9 A0A154PC81 A0A194QRC6 A0A2S2R4Y7 E2A711 F4W473 A0A195ETI6 E9I9W7 A0A212F0G1 A0A158NP49 W5JA84 A0A195CLI9 A0A067R8Z3 A0A023F7A3 E2A712 A0A195E4S8 A0A310SGK5 A0A069DY58 A0A182GQQ8 A0A182FDK1 A0A088A501 A0A2A3EAR6 A0A195CI97 J9HJE4
Q1HPX6 A0A194R949 H9J8I9 A0A194R584 H9J369 A0A0L7KQW0 A0A0H4TDA9 A0A194R4F0 H9J8I4 A0A2H1VNC8 A0A0F6Q2J9 A0A0L7LJP6 A0A2W1BR60 A0A0F6Q0W0 H9J8I5 A0A1L8D6N5 A0A212F936 A0A194QJQ6 A0A2W1BL60 A0A2A4JIR0 A0A194R3L5 A0A2W1BSK9 A0A0F6Q0X5 S4PIB4 H9J355 U5KC20 I4DKW3 A0A0H4TH26 D2SNK3 A0A212F911 H9J8I6 A0A0H4TD16 A0A0F6Q4H7 A0A2H1VSR5 A0A0L7K2F9 H9J354 A0A2H1VR53 A0A2A4JJW8 H9J353 H9J0A6 A0A2H8TFE7 A0A2S2NV02 J9JKN0 F4W474 A0A0C9R6P4 A0A026WTY5 A0A151WLU8 K7IV86 A0A232EGH1 A0A2P8Z2R9 A0A1W4WBZ1 A0A195ET92 A0A026WRA5 A0A0J7L0X1 Q6RUR3 A0A067QWG1 A0A151X1L8 E2B6N3 A0A0L7QSX6 A0A0J7L5T5 A0A140KPR8 B0XI43 A0A195B3G6 A0A195B441 A0A212F0F9 A0A154PC81 A0A194QRC6 A0A2S2R4Y7 E2A711 F4W473 A0A195ETI6 E9I9W7 A0A212F0G1 A0A158NP49 W5JA84 A0A195CLI9 A0A067R8Z3 A0A023F7A3 E2A712 A0A195E4S8 A0A310SGK5 A0A069DY58 A0A182GQQ8 A0A182FDK1 A0A088A501 A0A2A3EAR6 A0A195CI97 J9HJE4
Pubmed
EMBL
BABH01040078
BABH01043465
BABH01044234
BABH01043464
BABH01027538
DQ443276
+ More
ABF51365.1 KQ460779 KPJ12396.1 KPJ12400.1 BABH01036649 JTDY01007189 KOB65394.1 KP899549 AKQ06148.1 KPJ12399.1 BABH01027525 ODYU01003471 SOQ42306.1 KP237873 AKD01726.1 JTDY01000876 KOB75589.1 KZ149989 PZC75627.1 KP237884 AKD01737.1 BABH01027529 BABH01027530 GEYN01000013 JAV02116.1 AGBW02009651 OWR50237.1 KQ459144 KPJ03686.1 PZC75628.1 NWSH01001322 PCG71699.1 KPJ12398.1 PZC75626.1 KP237899 AKD01752.1 GAIX01005455 JAA87105.1 BABH01036642 KC007343 AGQ45608.1 AK401931 BAM18553.1 KPJ03685.1 KP899550 AKQ06149.1 EZ407131 ACX53694.1 OWR50236.1 BABH01027533 KP899551 AKQ06150.1 KP237875 AKD01728.1 ODYU01003911 SOQ43254.1 JTDY01017041 KOB51868.1 SOQ43256.1 PCG71700.1 BABH01036648 BABH01019209 GFXV01001021 MBW12826.1 GGMR01008279 MBY20898.1 ABLF02032246 ABLF02032258 GL887491 EGI71070.1 GBYB01012004 JAG81771.1 KK107119 EZA58569.1 KQ982944 KYQ48859.1 AAZX01008191 NNAY01004765 OXU17447.1 PYGN01000223 PSN50783.1 KQ981979 KYN31470.1 QOIP01000009 EZA58570.1 RLU18591.1 LBMM01001364 KMQ96442.1 AY491503 AAR84629.1 KK852879 KDR14474.1 KQ982585 KYQ54299.1 GL446000 EFN88617.1 KQ414755 KOC61725.1 LBMM01000659 KMQ97859.1 LN794625 CEO43692.1 DS233256 EDS28983.1 KQ976625 KYM79031.1 KYM79032.1 AGBW02011082 OWR47238.1 KQ434870 KZC09506.1 KQ461175 KPJ07894.1 GGMS01015878 MBY85081.1 GL437252 EFN70743.1 EGI71069.1 KYN31471.1 GL761846 EFZ22724.1 OWR47236.1 ADTU01001531 ADMH02001698 ETN61367.1 KQ977642 KYN00939.1 KK852855 KDR14911.1 GBBI01001633 JAC17079.1 EFN70744.1 KQ979609 KYN20168.1 KQ771134 OAD52542.1 GBGD01002580 JAC86309.1 JXUM01081117 KQ563228 KXJ74247.1 KZ288303 PBC28808.1 KQ977720 KYN00433.1 CH477964 EJY58014.1
ABF51365.1 KQ460779 KPJ12396.1 KPJ12400.1 BABH01036649 JTDY01007189 KOB65394.1 KP899549 AKQ06148.1 KPJ12399.1 BABH01027525 ODYU01003471 SOQ42306.1 KP237873 AKD01726.1 JTDY01000876 KOB75589.1 KZ149989 PZC75627.1 KP237884 AKD01737.1 BABH01027529 BABH01027530 GEYN01000013 JAV02116.1 AGBW02009651 OWR50237.1 KQ459144 KPJ03686.1 PZC75628.1 NWSH01001322 PCG71699.1 KPJ12398.1 PZC75626.1 KP237899 AKD01752.1 GAIX01005455 JAA87105.1 BABH01036642 KC007343 AGQ45608.1 AK401931 BAM18553.1 KPJ03685.1 KP899550 AKQ06149.1 EZ407131 ACX53694.1 OWR50236.1 BABH01027533 KP899551 AKQ06150.1 KP237875 AKD01728.1 ODYU01003911 SOQ43254.1 JTDY01017041 KOB51868.1 SOQ43256.1 PCG71700.1 BABH01036648 BABH01019209 GFXV01001021 MBW12826.1 GGMR01008279 MBY20898.1 ABLF02032246 ABLF02032258 GL887491 EGI71070.1 GBYB01012004 JAG81771.1 KK107119 EZA58569.1 KQ982944 KYQ48859.1 AAZX01008191 NNAY01004765 OXU17447.1 PYGN01000223 PSN50783.1 KQ981979 KYN31470.1 QOIP01000009 EZA58570.1 RLU18591.1 LBMM01001364 KMQ96442.1 AY491503 AAR84629.1 KK852879 KDR14474.1 KQ982585 KYQ54299.1 GL446000 EFN88617.1 KQ414755 KOC61725.1 LBMM01000659 KMQ97859.1 LN794625 CEO43692.1 DS233256 EDS28983.1 KQ976625 KYM79031.1 KYM79032.1 AGBW02011082 OWR47238.1 KQ434870 KZC09506.1 KQ461175 KPJ07894.1 GGMS01015878 MBY85081.1 GL437252 EFN70743.1 EGI71069.1 KYN31471.1 GL761846 EFZ22724.1 OWR47236.1 ADTU01001531 ADMH02001698 ETN61367.1 KQ977642 KYN00939.1 KK852855 KDR14911.1 GBBI01001633 JAC17079.1 EFN70744.1 KQ979609 KYN20168.1 KQ771134 OAD52542.1 GBGD01002580 JAC86309.1 JXUM01081117 KQ563228 KXJ74247.1 KZ288303 PBC28808.1 KQ977720 KYN00433.1 CH477964 EJY58014.1
Proteomes
UP000005204
UP000053240
UP000037510
UP000007151
UP000053268
UP000218220
+ More
UP000007819 UP000007755 UP000053097 UP000075809 UP000002358 UP000215335 UP000245037 UP000192223 UP000078541 UP000279307 UP000036403 UP000027135 UP000008237 UP000053825 UP000002320 UP000078540 UP000076502 UP000000311 UP000005205 UP000000673 UP000078542 UP000078492 UP000069940 UP000249989 UP000069272 UP000005203 UP000242457 UP000008820
UP000007819 UP000007755 UP000053097 UP000075809 UP000002358 UP000215335 UP000245037 UP000192223 UP000078541 UP000279307 UP000036403 UP000027135 UP000008237 UP000053825 UP000002320 UP000078540 UP000076502 UP000000311 UP000005205 UP000000673 UP000078542 UP000078492 UP000069940 UP000249989 UP000069272 UP000005203 UP000242457 UP000008820
PRIDE
Pfam
PF00106 adh_short
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
H9JWW7
H9JWW0
H9JYK0
H9JWW1
H9JWW6
H9J8J0
+ More
Q1HPX6 A0A194R949 H9J8I9 A0A194R584 H9J369 A0A0L7KQW0 A0A0H4TDA9 A0A194R4F0 H9J8I4 A0A2H1VNC8 A0A0F6Q2J9 A0A0L7LJP6 A0A2W1BR60 A0A0F6Q0W0 H9J8I5 A0A1L8D6N5 A0A212F936 A0A194QJQ6 A0A2W1BL60 A0A2A4JIR0 A0A194R3L5 A0A2W1BSK9 A0A0F6Q0X5 S4PIB4 H9J355 U5KC20 I4DKW3 A0A0H4TH26 D2SNK3 A0A212F911 H9J8I6 A0A0H4TD16 A0A0F6Q4H7 A0A2H1VSR5 A0A0L7K2F9 H9J354 A0A2H1VR53 A0A2A4JJW8 H9J353 H9J0A6 A0A2H8TFE7 A0A2S2NV02 J9JKN0 F4W474 A0A0C9R6P4 A0A026WTY5 A0A151WLU8 K7IV86 A0A232EGH1 A0A2P8Z2R9 A0A1W4WBZ1 A0A195ET92 A0A026WRA5 A0A0J7L0X1 Q6RUR3 A0A067QWG1 A0A151X1L8 E2B6N3 A0A0L7QSX6 A0A0J7L5T5 A0A140KPR8 B0XI43 A0A195B3G6 A0A195B441 A0A212F0F9 A0A154PC81 A0A194QRC6 A0A2S2R4Y7 E2A711 F4W473 A0A195ETI6 E9I9W7 A0A212F0G1 A0A158NP49 W5JA84 A0A195CLI9 A0A067R8Z3 A0A023F7A3 E2A712 A0A195E4S8 A0A310SGK5 A0A069DY58 A0A182GQQ8 A0A182FDK1 A0A088A501 A0A2A3EAR6 A0A195CI97 J9HJE4
Q1HPX6 A0A194R949 H9J8I9 A0A194R584 H9J369 A0A0L7KQW0 A0A0H4TDA9 A0A194R4F0 H9J8I4 A0A2H1VNC8 A0A0F6Q2J9 A0A0L7LJP6 A0A2W1BR60 A0A0F6Q0W0 H9J8I5 A0A1L8D6N5 A0A212F936 A0A194QJQ6 A0A2W1BL60 A0A2A4JIR0 A0A194R3L5 A0A2W1BSK9 A0A0F6Q0X5 S4PIB4 H9J355 U5KC20 I4DKW3 A0A0H4TH26 D2SNK3 A0A212F911 H9J8I6 A0A0H4TD16 A0A0F6Q4H7 A0A2H1VSR5 A0A0L7K2F9 H9J354 A0A2H1VR53 A0A2A4JJW8 H9J353 H9J0A6 A0A2H8TFE7 A0A2S2NV02 J9JKN0 F4W474 A0A0C9R6P4 A0A026WTY5 A0A151WLU8 K7IV86 A0A232EGH1 A0A2P8Z2R9 A0A1W4WBZ1 A0A195ET92 A0A026WRA5 A0A0J7L0X1 Q6RUR3 A0A067QWG1 A0A151X1L8 E2B6N3 A0A0L7QSX6 A0A0J7L5T5 A0A140KPR8 B0XI43 A0A195B3G6 A0A195B441 A0A212F0F9 A0A154PC81 A0A194QRC6 A0A2S2R4Y7 E2A711 F4W473 A0A195ETI6 E9I9W7 A0A212F0G1 A0A158NP49 W5JA84 A0A195CLI9 A0A067R8Z3 A0A023F7A3 E2A712 A0A195E4S8 A0A310SGK5 A0A069DY58 A0A182GQQ8 A0A182FDK1 A0A088A501 A0A2A3EAR6 A0A195CI97 J9HJE4
PDB
2GDZ
E-value=3.511e-23,
Score=266
Ontologies
GO
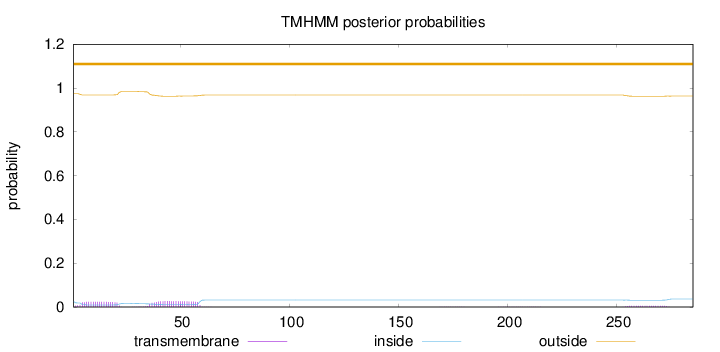
Topology
SignalP
Position: 1 - 18,
Likelihood: 0.980633
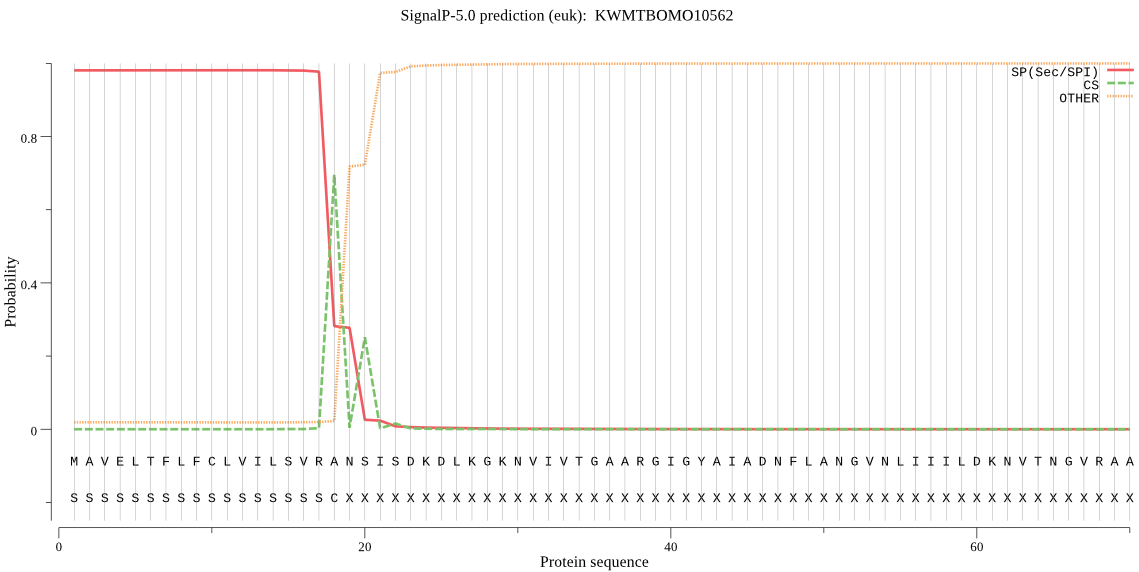
Length:
285
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.11483
Exp number, first 60 AAs:
0.99578
Total prob of N-in:
0.02551
outside
1 - 285
Population Genetic Test Statistics
Pi
157.36711
Theta
142.633354
Tajima's D
0.260816
CLR
0
CSRT
0.44402779861007
Interpretation
Uncertain
