Gene
KWMTBOMO10558 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007144
Annotation
PREDICTED:_3-hydroxyisobutyrate_dehydrogenase_isoform_X1_[Bombyx_mori]
Full name
3-hydroxyisobutyrate dehydrogenase
Location in the cell
Mitochondrial Reliability : 2.507
Sequence
CDS
ATGGCCGCGCGCACCATATTAAGCACTCAGTGTTTGTACACAGCCGCCAGACGGGCGTATAGTTCTAACACCGACAAGAATGTGGCTTTCCTCGGCCTCGGAAACATGGGAGGGTTCATGGCTGCGAACTTGGTTAAAAAGGGTTTCACAGTTCGCGGCTACGATCCTTCCAAAGATGCACTGAACGCAGCGGCCAAGAACGGAGTGACGCCAGCCAACTCCATTGCTGCAGCCGTTGATGGAGTCGACGTGGTCGTTTCCATATTGACCAGCAATAAAGTCGTGCTCGACGTGTACCTGGGCAAAGATGGCGTTGTGGCTCATGCGAAAAAAGGATCGCTTCTGATCGATTCGAGTACAATAGATCCGAATGTTCCCAAACAGATCTTCCCTATAGCCCTAGAGAAAGGGCTGGGATTCACAGATGCACCTGTATCTGGAGGAGTCATGGGTGCTCAGAACGCTACCCTGGCCTTCATGGCCGGAGGTCGTAAAGAAGACTTCGAAAGGTCCCTTCCTCTCCTCAAAGTGATGGGAGCTAAGCAGTTCCACTGCGGACAGATCGGCTCAGGACAGGTGGCTAAACTGACCAATAACATGTTGATGGGCATCACCGGAATGGCTACAGCTGAGTGCATGAACATGGGCATTAAAATGGGACTCGAACCGAAGGTTCTGTTGGACGTGCTGAACAACTCATCGGCTCGCTCCTGGTCGACCGAGGTGTACTGTCCAGTCCCCGGACTGGTGCCCACCGCTCCGTCGAGTAGAAATTACGATGGCGGCTTCAAAAACGAACTTATGGTTAAGGACTTAGAATTAGCCAGTGGCATGGCGTTGGGTATTCGTTCGCCGATCCCATTGGGCGCTGTCGCAACTCAGCTCTACCGCATCGTGCAGTCCAGGGGCTACGGCCAGAAAGACTTCTCGTTCGTCTTCCAGCTACTGAAAGAAGAAAACAAAAATTAG
Protein
MAARTILSTQCLYTAARRAYSSNTDKNVAFLGLGNMGGFMAANLVKKGFTVRGYDPSKDALNAAAKNGVTPANSIAAAVDGVDVVVSILTSNKVVLDVYLGKDGVVAHAKKGSLLIDSSTIDPNVPKQIFPIALEKGLGFTDAPVSGGVMGAQNATLAFMAGGRKEDFERSLPLLKVMGAKQFHCGQIGSGQVAKLTNNMLMGITGMATAECMNMGIKMGLEPKVLLDVLNNSSARSWSTEVYCPVPGLVPTAPSSRNYDGGFKNELMVKDLELASGMALGIRSPIPLGAVATQLYRIVQSRGYGQKDFSFVFQLLKEENKN
Summary
Catalytic Activity
3-hydroxy-2-methylpropanoate + NAD(+) = 2-methyl-3-oxopropanoate + H(+) + NADH
Similarity
Belongs to the HIBADH-related family.
Uniprot
H9JC99
B3GQU6
A0A2H1V253
A0A194RJP5
A0A2A4K6B3
A0A194PJC7
+ More
S4P4G9 I4DR58 A0A212F4X2 A0A232F4U2 E2BKV7 A0A026WLY7 A0A151X333 A0A1A9V4T6 A0A151I2H3 A0A1A9XAS1 A0A1A9ZX23 A0A0P5CP75 A0A0N8A5N0 A0A158NSB4 A0A0P5T8P1 A0A0P5BK54 A0A1B0G2W9 W8B751 D3TQ47 A0A0P5HKP8 A0A369RZ39 A0A1L8E147 A0A0A1X062 A0A1A9WNV5 A0A195ERZ3 A0A0P5HR85 A0A034VRZ1 E9GCN3 A0A0K8WJS2 A0A2A3EI45 A0A0K8VJQ9 A0A2P2HY37 A0A1S3JB31 A0A146P678 A0A1L8E1I8 V5I8T8 A0A1W7RFT5 A0A0F7Z8R5 T1DKE8 A0A098LZM6 A0A0S7HGG7 A0A3P9MXD6 T1DN42 A0A0B6YUP5 A0A3B4T5Q7 A0A315WD32 A0A2M3Z1E6 A0A3B5R058 A0A3Q2XI03 A0A2M4ABV3 A0A3B4XQP0 E2A2W8 A0A1U7S972 A0A3B5QWB3 H2UQQ7 A0A3Q3LQE4 A0A3Q1HD37 A0A3B4GAF1 A0A3Q3IBG0 A0A3Q3M485 A0A3P8NKB1 A0A3P9C8X0 A8DSV9 A0A151MR22 A0A2M4BTZ9 A0A2D0RUI2 L8HDI8 A0A2M3Z1N8 A0A3Q2E582 A0A0S7HE98 A0A182LJB1 A0A3B3XSR0 A0A3B3DSE3 A0A3P9I6H8 A0A2M3Z1J6 A0A3Q3VSV9 W5J8L6 C3Y133 A0A3P9K560 A0A182PHH0 A0A182FEI1 A0A0F8ADE7 A0A3Q3JGK6 Q1KKZ7 A0A3S2P847 A0A1B6GZS7 A0A3P8V030 A0A3B5KQ40 H2LTU1 A0A3P9IJU4 A0A182VGD6 A0A2M4BTL5 A0A3P9L0W1
S4P4G9 I4DR58 A0A212F4X2 A0A232F4U2 E2BKV7 A0A026WLY7 A0A151X333 A0A1A9V4T6 A0A151I2H3 A0A1A9XAS1 A0A1A9ZX23 A0A0P5CP75 A0A0N8A5N0 A0A158NSB4 A0A0P5T8P1 A0A0P5BK54 A0A1B0G2W9 W8B751 D3TQ47 A0A0P5HKP8 A0A369RZ39 A0A1L8E147 A0A0A1X062 A0A1A9WNV5 A0A195ERZ3 A0A0P5HR85 A0A034VRZ1 E9GCN3 A0A0K8WJS2 A0A2A3EI45 A0A0K8VJQ9 A0A2P2HY37 A0A1S3JB31 A0A146P678 A0A1L8E1I8 V5I8T8 A0A1W7RFT5 A0A0F7Z8R5 T1DKE8 A0A098LZM6 A0A0S7HGG7 A0A3P9MXD6 T1DN42 A0A0B6YUP5 A0A3B4T5Q7 A0A315WD32 A0A2M3Z1E6 A0A3B5R058 A0A3Q2XI03 A0A2M4ABV3 A0A3B4XQP0 E2A2W8 A0A1U7S972 A0A3B5QWB3 H2UQQ7 A0A3Q3LQE4 A0A3Q1HD37 A0A3B4GAF1 A0A3Q3IBG0 A0A3Q3M485 A0A3P8NKB1 A0A3P9C8X0 A8DSV9 A0A151MR22 A0A2M4BTZ9 A0A2D0RUI2 L8HDI8 A0A2M3Z1N8 A0A3Q2E582 A0A0S7HE98 A0A182LJB1 A0A3B3XSR0 A0A3B3DSE3 A0A3P9I6H8 A0A2M3Z1J6 A0A3Q3VSV9 W5J8L6 C3Y133 A0A3P9K560 A0A182PHH0 A0A182FEI1 A0A0F8ADE7 A0A3Q3JGK6 Q1KKZ7 A0A3S2P847 A0A1B6GZS7 A0A3P8V030 A0A3B5KQ40 H2LTU1 A0A3P9IJU4 A0A182VGD6 A0A2M4BTL5 A0A3P9L0W1
EC Number
1.1.1.31
Pubmed
19121390
26354079
23622113
22651552
22118469
28648823
+ More
20798317 24508170 21347285 24495485 20353571 30042472 25830018 25348373 21292972 26383154 26358130 23758969 25727380 29703783 23542700 21551351 25186727 17845724 22293439 23375108 20966253 29451363 17554307 20920257 23761445 18563158 25835551 16636282 24487278
20798317 24508170 21347285 24495485 20353571 30042472 25830018 25348373 21292972 26383154 26358130 23758969 25727380 29703783 23542700 21551351 25186727 17845724 22293439 23375108 20966253 29451363 17554307 20920257 23761445 18563158 25835551 16636282 24487278
EMBL
BABH01015134
EU719652
ACD92999.1
ODYU01000337
SOQ34911.1
KQ460124
+ More
KPJ17659.1 NWSH01000133 PCG79210.1 KQ459603 KPI93143.1 GAIX01005969 JAA86591.1 AK404956 BAM20398.1 AGBW02010289 OWR48798.1 NNAY01000942 OXU25801.1 GL448826 EFN83706.1 KK107171 EZA56099.1 KQ982562 KYQ54835.1 KQ976543 KYM81008.1 GDIP01183447 JAJ39955.1 GDIP01180097 JAJ43305.1 ADTU01024780 GDIP01217791 GDIP01213050 GDIQ01241264 GDIQ01208968 GDIQ01154185 GDIQ01138344 GDIP01131661 GDIP01094509 JAK10461.1 JAL72053.1 GDIP01183448 JAJ39954.1 CCAG010015288 GAMC01013642 GAMC01013641 JAB92913.1 EZ423549 ADD19825.1 GDIQ01231713 GDIQ01121050 JAK20012.1 NOWV01000116 RDD39948.1 GFDF01001718 JAV12366.1 GBXI01010127 JAD04165.1 KQ981993 KYN30998.1 GDIQ01229461 JAK22264.1 GAKP01014065 JAC44887.1 GL732539 EFX82586.1 GDHF01027621 GDHF01022291 GDHF01012221 GDHF01004573 GDHF01001020 JAI24693.1 JAI30023.1 JAI40093.1 JAI47741.1 JAI51294.1 KZ288253 PBC30942.1 GDHF01013190 JAI39124.1 IACF01000960 LAB66694.1 GCES01147306 GCES01146523 JAQ39016.1 GFDF01001719 JAV12365.1 GALX01004110 JAB64356.1 GDAY02001414 JAV50013.1 GBEX01001987 JAI12573.1 GAAZ01001418 GBKC01002103 GBKD01001263 JAA96525.1 JAG43967.1 JAG46355.1 GBSI01001072 JAC95424.1 GBYX01442853 JAO38541.1 GAMD01003205 JAA98385.1 HACG01012962 CEK59827.1 NHOQ01000034 PWA33647.1 GGFM01001560 MBW22311.1 GGFK01004767 MBW38088.1 GL436229 EFN72210.1 EF594311 ABS70747.1 AKHW03005461 KYO26850.1 GGFJ01007280 MBW56421.1 KB007857 ELR23250.1 GGFM01001672 MBW22423.1 GBYX01442852 JAO38542.1 GGFM01001639 MBW22390.1 ADMH02002097 ETN59220.1 GG666480 EEN65573.1 KQ042840 KKF09948.1 DQ481664 ABF22397.1 CM012447 RVE66853.1 GECZ01001847 JAS67922.1 GGFJ01007276 MBW56417.1
KPJ17659.1 NWSH01000133 PCG79210.1 KQ459603 KPI93143.1 GAIX01005969 JAA86591.1 AK404956 BAM20398.1 AGBW02010289 OWR48798.1 NNAY01000942 OXU25801.1 GL448826 EFN83706.1 KK107171 EZA56099.1 KQ982562 KYQ54835.1 KQ976543 KYM81008.1 GDIP01183447 JAJ39955.1 GDIP01180097 JAJ43305.1 ADTU01024780 GDIP01217791 GDIP01213050 GDIQ01241264 GDIQ01208968 GDIQ01154185 GDIQ01138344 GDIP01131661 GDIP01094509 JAK10461.1 JAL72053.1 GDIP01183448 JAJ39954.1 CCAG010015288 GAMC01013642 GAMC01013641 JAB92913.1 EZ423549 ADD19825.1 GDIQ01231713 GDIQ01121050 JAK20012.1 NOWV01000116 RDD39948.1 GFDF01001718 JAV12366.1 GBXI01010127 JAD04165.1 KQ981993 KYN30998.1 GDIQ01229461 JAK22264.1 GAKP01014065 JAC44887.1 GL732539 EFX82586.1 GDHF01027621 GDHF01022291 GDHF01012221 GDHF01004573 GDHF01001020 JAI24693.1 JAI30023.1 JAI40093.1 JAI47741.1 JAI51294.1 KZ288253 PBC30942.1 GDHF01013190 JAI39124.1 IACF01000960 LAB66694.1 GCES01147306 GCES01146523 JAQ39016.1 GFDF01001719 JAV12365.1 GALX01004110 JAB64356.1 GDAY02001414 JAV50013.1 GBEX01001987 JAI12573.1 GAAZ01001418 GBKC01002103 GBKD01001263 JAA96525.1 JAG43967.1 JAG46355.1 GBSI01001072 JAC95424.1 GBYX01442853 JAO38541.1 GAMD01003205 JAA98385.1 HACG01012962 CEK59827.1 NHOQ01000034 PWA33647.1 GGFM01001560 MBW22311.1 GGFK01004767 MBW38088.1 GL436229 EFN72210.1 EF594311 ABS70747.1 AKHW03005461 KYO26850.1 GGFJ01007280 MBW56421.1 KB007857 ELR23250.1 GGFM01001672 MBW22423.1 GBYX01442852 JAO38542.1 GGFM01001639 MBW22390.1 ADMH02002097 ETN59220.1 GG666480 EEN65573.1 KQ042840 KKF09948.1 DQ481664 ABF22397.1 CM012447 RVE66853.1 GECZ01001847 JAS67922.1 GGFJ01007276 MBW56417.1
Proteomes
UP000005204
UP000053240
UP000218220
UP000053268
UP000007151
UP000215335
+ More
UP000008237 UP000053097 UP000075809 UP000078200 UP000078540 UP000092443 UP000092445 UP000005205 UP000092444 UP000253843 UP000091820 UP000078541 UP000000305 UP000242457 UP000085678 UP000265000 UP000242638 UP000261420 UP000002852 UP000264820 UP000261360 UP000000311 UP000189705 UP000005226 UP000261640 UP000265040 UP000261460 UP000261600 UP000265100 UP000265160 UP000264840 UP000050525 UP000221080 UP000011083 UP000265020 UP000075882 UP000261480 UP000261560 UP000265200 UP000261620 UP000000673 UP000001554 UP000265180 UP000075885 UP000069272 UP000265120 UP000001038 UP000075903
UP000008237 UP000053097 UP000075809 UP000078200 UP000078540 UP000092443 UP000092445 UP000005205 UP000092444 UP000253843 UP000091820 UP000078541 UP000000305 UP000242457 UP000085678 UP000265000 UP000242638 UP000261420 UP000002852 UP000264820 UP000261360 UP000000311 UP000189705 UP000005226 UP000261640 UP000265040 UP000261460 UP000261600 UP000265100 UP000265160 UP000264840 UP000050525 UP000221080 UP000011083 UP000265020 UP000075882 UP000261480 UP000261560 UP000265200 UP000261620 UP000000673 UP000001554 UP000265180 UP000075885 UP000069272 UP000265120 UP000001038 UP000075903
Interpro
Gene 3D
ProteinModelPortal
H9JC99
B3GQU6
A0A2H1V253
A0A194RJP5
A0A2A4K6B3
A0A194PJC7
+ More
S4P4G9 I4DR58 A0A212F4X2 A0A232F4U2 E2BKV7 A0A026WLY7 A0A151X333 A0A1A9V4T6 A0A151I2H3 A0A1A9XAS1 A0A1A9ZX23 A0A0P5CP75 A0A0N8A5N0 A0A158NSB4 A0A0P5T8P1 A0A0P5BK54 A0A1B0G2W9 W8B751 D3TQ47 A0A0P5HKP8 A0A369RZ39 A0A1L8E147 A0A0A1X062 A0A1A9WNV5 A0A195ERZ3 A0A0P5HR85 A0A034VRZ1 E9GCN3 A0A0K8WJS2 A0A2A3EI45 A0A0K8VJQ9 A0A2P2HY37 A0A1S3JB31 A0A146P678 A0A1L8E1I8 V5I8T8 A0A1W7RFT5 A0A0F7Z8R5 T1DKE8 A0A098LZM6 A0A0S7HGG7 A0A3P9MXD6 T1DN42 A0A0B6YUP5 A0A3B4T5Q7 A0A315WD32 A0A2M3Z1E6 A0A3B5R058 A0A3Q2XI03 A0A2M4ABV3 A0A3B4XQP0 E2A2W8 A0A1U7S972 A0A3B5QWB3 H2UQQ7 A0A3Q3LQE4 A0A3Q1HD37 A0A3B4GAF1 A0A3Q3IBG0 A0A3Q3M485 A0A3P8NKB1 A0A3P9C8X0 A8DSV9 A0A151MR22 A0A2M4BTZ9 A0A2D0RUI2 L8HDI8 A0A2M3Z1N8 A0A3Q2E582 A0A0S7HE98 A0A182LJB1 A0A3B3XSR0 A0A3B3DSE3 A0A3P9I6H8 A0A2M3Z1J6 A0A3Q3VSV9 W5J8L6 C3Y133 A0A3P9K560 A0A182PHH0 A0A182FEI1 A0A0F8ADE7 A0A3Q3JGK6 Q1KKZ7 A0A3S2P847 A0A1B6GZS7 A0A3P8V030 A0A3B5KQ40 H2LTU1 A0A3P9IJU4 A0A182VGD6 A0A2M4BTL5 A0A3P9L0W1
S4P4G9 I4DR58 A0A212F4X2 A0A232F4U2 E2BKV7 A0A026WLY7 A0A151X333 A0A1A9V4T6 A0A151I2H3 A0A1A9XAS1 A0A1A9ZX23 A0A0P5CP75 A0A0N8A5N0 A0A158NSB4 A0A0P5T8P1 A0A0P5BK54 A0A1B0G2W9 W8B751 D3TQ47 A0A0P5HKP8 A0A369RZ39 A0A1L8E147 A0A0A1X062 A0A1A9WNV5 A0A195ERZ3 A0A0P5HR85 A0A034VRZ1 E9GCN3 A0A0K8WJS2 A0A2A3EI45 A0A0K8VJQ9 A0A2P2HY37 A0A1S3JB31 A0A146P678 A0A1L8E1I8 V5I8T8 A0A1W7RFT5 A0A0F7Z8R5 T1DKE8 A0A098LZM6 A0A0S7HGG7 A0A3P9MXD6 T1DN42 A0A0B6YUP5 A0A3B4T5Q7 A0A315WD32 A0A2M3Z1E6 A0A3B5R058 A0A3Q2XI03 A0A2M4ABV3 A0A3B4XQP0 E2A2W8 A0A1U7S972 A0A3B5QWB3 H2UQQ7 A0A3Q3LQE4 A0A3Q1HD37 A0A3B4GAF1 A0A3Q3IBG0 A0A3Q3M485 A0A3P8NKB1 A0A3P9C8X0 A8DSV9 A0A151MR22 A0A2M4BTZ9 A0A2D0RUI2 L8HDI8 A0A2M3Z1N8 A0A3Q2E582 A0A0S7HE98 A0A182LJB1 A0A3B3XSR0 A0A3B3DSE3 A0A3P9I6H8 A0A2M3Z1J6 A0A3Q3VSV9 W5J8L6 C3Y133 A0A3P9K560 A0A182PHH0 A0A182FEI1 A0A0F8ADE7 A0A3Q3JGK6 Q1KKZ7 A0A3S2P847 A0A1B6GZS7 A0A3P8V030 A0A3B5KQ40 H2LTU1 A0A3P9IJU4 A0A182VGD6 A0A2M4BTL5 A0A3P9L0W1
PDB
2I9P
E-value=3.8415e-65,
Score=629
Ontologies
PATHWAY
00280
Valine, leucine and isoleucine degradation - Bombyx mori (domestic silkworm)
00410 beta-Alanine metabolism - Bombyx mori (domestic silkworm)
00640 Propanoate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
00410 beta-Alanine metabolism - Bombyx mori (domestic silkworm)
00640 Propanoate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
GO
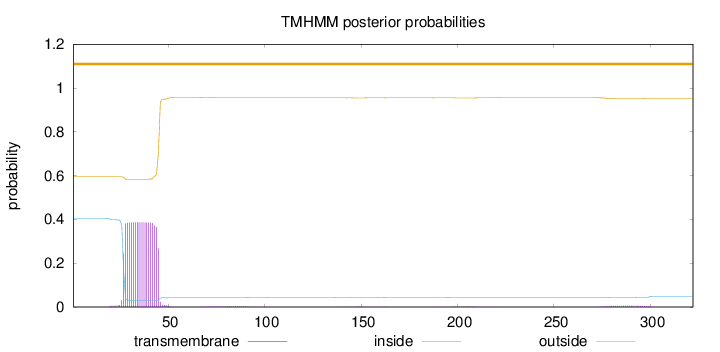
Topology
Length:
322
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
7.40525
Exp number, first 60 AAs:
7.15559
Total prob of N-in:
0.40494
outside
1 - 322
Population Genetic Test Statistics
Pi
245.552836
Theta
149.197322
Tajima's D
1.748495
CLR
0.348776
CSRT
0.84050797460127
Interpretation
Uncertain
