Gene
KWMTBOMO10557
Pre Gene Modal
BGIBMGA007143
Annotation
PREDICTED:_sulfotransferase_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.563 Mitochondrial Reliability : 1.212
Sequence
CDS
ATGCCAGCCAAGATGGGTCCTCAAATCGTTCTGCCATACGACATAGAAGACGTCACGGCAGACGAGGATAAAATAATAAAAAGGTGCTTATTAGGCTACACCAGACCATTCGTGAAATGCGGTAAGAAGGGCTACGTGATGCCGGGCGCGTTCAGGAAACACGCAGATGCCATCTACAACATGAAAGTTAGACCCGATGATATTTGGGTCATCACTTTCCCGAGATCAGGTACAACGTGGACGCAAGAATTAATTTGGCTGGTGGAAAATAATTTGAATTATAACGCTGCAAGAGAACGACCCCTTTACGAAAGATTTCCAATGCTAGAAACAACCAGCCAGATACCGGAAATAGCGTACGAGTTCATAAAAGCGAATTTCATGAATTTGGGCAGCTTCCAAGGGCTTACGGAAGCGGCCCGGTACCCTAGCTGGAAGATGATAGAGGAGATGCCTTCCCCGAGATTCATAAAGACACACCTGCCACTATCCCTTCTGCCGCCGACCCTGCTGGAGACGGCAAAGGTCGTGTACGTGGCCAGGGATCCAAGGGATGTTGTGGTTTCATATTATTATCTACACAAAATGGTCAGCAAGCATTTGATGAGGGCTACGTTTCACGACTTCTGGGAGGCCTTCAGACGAGATCTGTTACCTTGGACCCCAATAGTAGCACACACAAACGAGGCCTGGGTCCATCGATACCATCCGAACATGCACTTTGTGTTTTATGAAGATATGATTAAGGATCTGCCCAAGGAAATCCGTCGAGTCTGTAAGTTCTTGCAGAGGTACTACACGGACGACCAAGTGAAGGCGCTCTCAGAGCATCTCAGTTTTGAGAGCTTAAGAAAAAACAAGAATGTTAACAATACTACCGGCGAAGAGAATAATGGAGTGCAGTTTGTAAGGAAAGGTGAAGCCGGTGGTTGGAAGTCGCATTTTGACGAAGTGATGACGTCACAAGCCGAAGACTTCCTCATATCGCGACTGAAAGGTCTAGATCTCGTGTACCCGTCGTTGGAAAACGGGGAGACCACACATCTATAG
Protein
MPAKMGPQIVLPYDIEDVTADEDKIIKRCLLGYTRPFVKCGKKGYVMPGAFRKHADAIYNMKVRPDDIWVITFPRSGTTWTQELIWLVENNLNYNAARERPLYERFPMLETTSQIPEIAYEFIKANFMNLGSFQGLTEAARYPSWKMIEEMPSPRFIKTHLPLSLLPPTLLETAKVVYVARDPRDVVVSYYYLHKMVSKHLMRATFHDFWEAFRRDLLPWTPIVAHTNEAWVHRYHPNMHFVFYEDMIKDLPKEIRRVCKFLQRYYTDDQVKALSEHLSFESLRKNKNVNNTTGEENNGVQFVRKGEAGGWKSHFDEVMTSQAEDFLISRLKGLDLVYPSLENGETTHL
Summary
Uniprot
H9JC98
Q3LFP9
A0A2A4K568
A0A1E1W8N3
A0A194RJZ8
A0A194PK88
+ More
A0A2H1V3G5 A0A212FHR1 E3UKQ3 A0A2H1VMK2 A0A0L7LHU2 A0A2A4K5L1 H9JBS7 A0PCF9 Q1HPJ5 A0A2H1WDE5 I4DJJ6 A0A194RNR7 A0A212FII0 A0A194PPT7 A0A194QMT8 H9JB37 A0A2H1V382 A0A194QI62 A0A0N1IHS9 A0A0L7QLZ9 A0A2H1X2F1 A0A3Q0JEY6 A0A3Q0JIS4 A0A2A3EI93 A0A2J7RQY3 A0A2H1WC36 A0A2H1VZQ2 A0A194QNL9 A0A3S2PJD2 A0A2A4KBH0 A0A3Q0JEW1 A0A1B6DDS5 A0A232F572 A0A2A4KAH9 K7IXC8 A0A088AQ03 A0A195C9J1 A0A0C9RP37 A0A212FIH7 F4W9H3 Q26490 A0A194QL22 A0A224XS10 A0A3S2M7E4 E2AHR9 A0A067QQ48 A0A0N1IET4 A0A1B0Y0B2 A0A194QHU7 A0A0A9VT74 A0A069DSX1 E9GDV9 A0A2H1WC50 A0A0V0G967 A0A224XSY7 A0A212FIG6 A0A151IWM2 A0A0V0G6A3 K7IVH8 A0A195CPN9 A0A224XN75 A0A194QHR0 A0A224XRN0 A0A0M8ZSK1 A0A069DSW0 E2BKD5 A0A151WNL9 A0A151IPX0 H9JBS6 A0A0P5BW27 A0A0P5BGU9 A0A0P5BKL0 E2BKD3 A0A0P5BS40 A0A195DUR6 A0A0P5G8H7 A0A0P5Y1N1 A0A0P5Z523 A0A0P5C3I9 A0A0N8D9T5 A0A224XU45 A0A0P5LRK9 A0A164ZQS0 A0A0P5SY28 A0A0N8BUL6 A0A224XTD2 A0A0P5RQL9 A0A0P6FPG4 A0A0P5N4Y8 A0A3L8E1L6 A0A069DSB1 A0A0P5BUH5 A0A0P6EDE3 A0A023F783
A0A2H1V3G5 A0A212FHR1 E3UKQ3 A0A2H1VMK2 A0A0L7LHU2 A0A2A4K5L1 H9JBS7 A0PCF9 Q1HPJ5 A0A2H1WDE5 I4DJJ6 A0A194RNR7 A0A212FII0 A0A194PPT7 A0A194QMT8 H9JB37 A0A2H1V382 A0A194QI62 A0A0N1IHS9 A0A0L7QLZ9 A0A2H1X2F1 A0A3Q0JEY6 A0A3Q0JIS4 A0A2A3EI93 A0A2J7RQY3 A0A2H1WC36 A0A2H1VZQ2 A0A194QNL9 A0A3S2PJD2 A0A2A4KBH0 A0A3Q0JEW1 A0A1B6DDS5 A0A232F572 A0A2A4KAH9 K7IXC8 A0A088AQ03 A0A195C9J1 A0A0C9RP37 A0A212FIH7 F4W9H3 Q26490 A0A194QL22 A0A224XS10 A0A3S2M7E4 E2AHR9 A0A067QQ48 A0A0N1IET4 A0A1B0Y0B2 A0A194QHU7 A0A0A9VT74 A0A069DSX1 E9GDV9 A0A2H1WC50 A0A0V0G967 A0A224XSY7 A0A212FIG6 A0A151IWM2 A0A0V0G6A3 K7IVH8 A0A195CPN9 A0A224XN75 A0A194QHR0 A0A224XRN0 A0A0M8ZSK1 A0A069DSW0 E2BKD5 A0A151WNL9 A0A151IPX0 H9JBS6 A0A0P5BW27 A0A0P5BGU9 A0A0P5BKL0 E2BKD3 A0A0P5BS40 A0A195DUR6 A0A0P5G8H7 A0A0P5Y1N1 A0A0P5Z523 A0A0P5C3I9 A0A0N8D9T5 A0A224XU45 A0A0P5LRK9 A0A164ZQS0 A0A0P5SY28 A0A0N8BUL6 A0A224XTD2 A0A0P5RQL9 A0A0P6FPG4 A0A0P5N4Y8 A0A3L8E1L6 A0A069DSB1 A0A0P5BUH5 A0A0P6EDE3 A0A023F783
Pubmed
EMBL
BABH01015133
AB237160
BAE46394.1
NWSH01000133
PCG79209.1
GDQN01007833
+ More
JAT83221.1 KQ460124 KPJ17660.1 KQ459603 KPI93144.1 ODYU01000337 SOQ34912.1 AGBW02008469 OWR53272.1 HM449927 ADO33062.1 ODYU01003367 SOQ42038.1 JTDY01001067 KOB74975.1 PCG79208.1 BABH01015119 BABH01015120 AB284983 BAF37537.1 DQ443407 ABF51496.1 ODYU01007908 SOQ51100.1 AK401464 BAM18086.1 KPJ17661.1 AGBW02008382 OWR53534.1 KPI93145.1 KQ461198 KPJ06275.1 BABH01008529 BABH01008530 BABH01008531 ODYU01000444 SOQ35226.1 KQ458793 KPJ05084.1 KQ460458 KPJ14654.1 KQ414905 KOC59625.1 ODYU01012890 SOQ59422.1 KZ288232 PBC31513.1 NEVH01000613 PNF43242.1 ODYU01007663 SOQ50641.1 ODYU01005448 SOQ46321.1 KPJ05081.1 RSAL01000015 RVE53120.1 NWSH01000001 PCG81090.1 GEDC01013553 JAS23745.1 NNAY01000980 OXU25629.1 PCG81089.1 AAZX01000018 KQ978068 KYM97385.1 GBYB01015302 JAG85069.1 OWR53536.1 GL888030 EGI69148.1 U28654 AAC47136.1 KPJ06273.1 GFTR01005206 JAW11220.1 RSAL01000014 RVE53184.1 GL439579 EFN66998.1 KK853064 KDR11875.1 LADJ01060126 KPJ21606.1 KT000403 ANI86001.1 KPJ05082.1 GBHO01044077 GBRD01004179 GDHC01002349 JAF99526.1 JAG61642.1 JAQ16280.1 GBGD01002073 JAC86816.1 GL732540 EFX82151.1 SOQ50640.1 GECL01001637 JAP04487.1 GFTR01005203 JAW11223.1 OWR53535.1 KQ980849 KYN12229.1 GECL01002505 JAP03619.1 KQ977444 KYN02703.1 GFTR01005158 JAW11268.1 KPJ05083.1 GFTR01005326 JAW11100.1 KQ435922 KOX68659.1 GBGD01002083 JAC86806.1 GL448783 EFN83859.1 KQ982907 KYQ49408.1 KQ976806 KYN08201.1 BABH01015125 GDIP01231190 GDIP01183245 JAJ40157.1 GDIP01184591 JAJ38811.1 GDIP01183247 JAJ40155.1 EFN83857.1 GDIP01183246 GDIP01181589 JAJ41813.1 KQ980322 KYN16645.1 GDIQ01244754 GDIQ01130895 JAK06971.1 GDIQ01162738 GDIP01068402 GDIP01064206 JAK88987.1 JAM39509.1 GDIP01152046 GDIP01049010 JAM54705.1 GDIP01176136 JAJ47266.1 GDIP01054767 JAM48948.1 GFTR01004803 JAW11623.1 GDIQ01168270 GDIQ01091393 JAK83455.1 LRGB01000687 KZS16634.1 GDIP01136443 JAL67271.1 GDIQ01267718 GDIQ01143331 GDIQ01109342 GDIQ01097394 JAL08395.1 GFTR01005205 JAW11221.1 GDIQ01097393 JAL54333.1 GDIQ01244755 GDIQ01129591 GDIQ01044578 JAN50159.1 GDIQ01169122 GDIQ01066475 JAK82603.1 QOIP01000001 RLU26620.1 GBGD01002074 JAC86815.1 GDIP01179738 JAJ43664.1 GDIQ01076601 JAN18136.1 GBBI01001612 JAC17100.1
JAT83221.1 KQ460124 KPJ17660.1 KQ459603 KPI93144.1 ODYU01000337 SOQ34912.1 AGBW02008469 OWR53272.1 HM449927 ADO33062.1 ODYU01003367 SOQ42038.1 JTDY01001067 KOB74975.1 PCG79208.1 BABH01015119 BABH01015120 AB284983 BAF37537.1 DQ443407 ABF51496.1 ODYU01007908 SOQ51100.1 AK401464 BAM18086.1 KPJ17661.1 AGBW02008382 OWR53534.1 KPI93145.1 KQ461198 KPJ06275.1 BABH01008529 BABH01008530 BABH01008531 ODYU01000444 SOQ35226.1 KQ458793 KPJ05084.1 KQ460458 KPJ14654.1 KQ414905 KOC59625.1 ODYU01012890 SOQ59422.1 KZ288232 PBC31513.1 NEVH01000613 PNF43242.1 ODYU01007663 SOQ50641.1 ODYU01005448 SOQ46321.1 KPJ05081.1 RSAL01000015 RVE53120.1 NWSH01000001 PCG81090.1 GEDC01013553 JAS23745.1 NNAY01000980 OXU25629.1 PCG81089.1 AAZX01000018 KQ978068 KYM97385.1 GBYB01015302 JAG85069.1 OWR53536.1 GL888030 EGI69148.1 U28654 AAC47136.1 KPJ06273.1 GFTR01005206 JAW11220.1 RSAL01000014 RVE53184.1 GL439579 EFN66998.1 KK853064 KDR11875.1 LADJ01060126 KPJ21606.1 KT000403 ANI86001.1 KPJ05082.1 GBHO01044077 GBRD01004179 GDHC01002349 JAF99526.1 JAG61642.1 JAQ16280.1 GBGD01002073 JAC86816.1 GL732540 EFX82151.1 SOQ50640.1 GECL01001637 JAP04487.1 GFTR01005203 JAW11223.1 OWR53535.1 KQ980849 KYN12229.1 GECL01002505 JAP03619.1 KQ977444 KYN02703.1 GFTR01005158 JAW11268.1 KPJ05083.1 GFTR01005326 JAW11100.1 KQ435922 KOX68659.1 GBGD01002083 JAC86806.1 GL448783 EFN83859.1 KQ982907 KYQ49408.1 KQ976806 KYN08201.1 BABH01015125 GDIP01231190 GDIP01183245 JAJ40157.1 GDIP01184591 JAJ38811.1 GDIP01183247 JAJ40155.1 EFN83857.1 GDIP01183246 GDIP01181589 JAJ41813.1 KQ980322 KYN16645.1 GDIQ01244754 GDIQ01130895 JAK06971.1 GDIQ01162738 GDIP01068402 GDIP01064206 JAK88987.1 JAM39509.1 GDIP01152046 GDIP01049010 JAM54705.1 GDIP01176136 JAJ47266.1 GDIP01054767 JAM48948.1 GFTR01004803 JAW11623.1 GDIQ01168270 GDIQ01091393 JAK83455.1 LRGB01000687 KZS16634.1 GDIP01136443 JAL67271.1 GDIQ01267718 GDIQ01143331 GDIQ01109342 GDIQ01097394 JAL08395.1 GFTR01005205 JAW11221.1 GDIQ01097393 JAL54333.1 GDIQ01244755 GDIQ01129591 GDIQ01044578 JAN50159.1 GDIQ01169122 GDIQ01066475 JAK82603.1 QOIP01000001 RLU26620.1 GBGD01002074 JAC86815.1 GDIP01179738 JAJ43664.1 GDIQ01076601 JAN18136.1 GBBI01001612 JAC17100.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
+ More
UP000053825 UP000079169 UP000242457 UP000235965 UP000283053 UP000215335 UP000002358 UP000005203 UP000078542 UP000007755 UP000000311 UP000027135 UP000000305 UP000078492 UP000053105 UP000008237 UP000075809 UP000076858 UP000279307
UP000053825 UP000079169 UP000242457 UP000235965 UP000283053 UP000215335 UP000002358 UP000005203 UP000078542 UP000007755 UP000000311 UP000027135 UP000000305 UP000078492 UP000053105 UP000008237 UP000075809 UP000076858 UP000279307
Interpro
ProteinModelPortal
H9JC98
Q3LFP9
A0A2A4K568
A0A1E1W8N3
A0A194RJZ8
A0A194PK88
+ More
A0A2H1V3G5 A0A212FHR1 E3UKQ3 A0A2H1VMK2 A0A0L7LHU2 A0A2A4K5L1 H9JBS7 A0PCF9 Q1HPJ5 A0A2H1WDE5 I4DJJ6 A0A194RNR7 A0A212FII0 A0A194PPT7 A0A194QMT8 H9JB37 A0A2H1V382 A0A194QI62 A0A0N1IHS9 A0A0L7QLZ9 A0A2H1X2F1 A0A3Q0JEY6 A0A3Q0JIS4 A0A2A3EI93 A0A2J7RQY3 A0A2H1WC36 A0A2H1VZQ2 A0A194QNL9 A0A3S2PJD2 A0A2A4KBH0 A0A3Q0JEW1 A0A1B6DDS5 A0A232F572 A0A2A4KAH9 K7IXC8 A0A088AQ03 A0A195C9J1 A0A0C9RP37 A0A212FIH7 F4W9H3 Q26490 A0A194QL22 A0A224XS10 A0A3S2M7E4 E2AHR9 A0A067QQ48 A0A0N1IET4 A0A1B0Y0B2 A0A194QHU7 A0A0A9VT74 A0A069DSX1 E9GDV9 A0A2H1WC50 A0A0V0G967 A0A224XSY7 A0A212FIG6 A0A151IWM2 A0A0V0G6A3 K7IVH8 A0A195CPN9 A0A224XN75 A0A194QHR0 A0A224XRN0 A0A0M8ZSK1 A0A069DSW0 E2BKD5 A0A151WNL9 A0A151IPX0 H9JBS6 A0A0P5BW27 A0A0P5BGU9 A0A0P5BKL0 E2BKD3 A0A0P5BS40 A0A195DUR6 A0A0P5G8H7 A0A0P5Y1N1 A0A0P5Z523 A0A0P5C3I9 A0A0N8D9T5 A0A224XU45 A0A0P5LRK9 A0A164ZQS0 A0A0P5SY28 A0A0N8BUL6 A0A224XTD2 A0A0P5RQL9 A0A0P6FPG4 A0A0P5N4Y8 A0A3L8E1L6 A0A069DSB1 A0A0P5BUH5 A0A0P6EDE3 A0A023F783
A0A2H1V3G5 A0A212FHR1 E3UKQ3 A0A2H1VMK2 A0A0L7LHU2 A0A2A4K5L1 H9JBS7 A0PCF9 Q1HPJ5 A0A2H1WDE5 I4DJJ6 A0A194RNR7 A0A212FII0 A0A194PPT7 A0A194QMT8 H9JB37 A0A2H1V382 A0A194QI62 A0A0N1IHS9 A0A0L7QLZ9 A0A2H1X2F1 A0A3Q0JEY6 A0A3Q0JIS4 A0A2A3EI93 A0A2J7RQY3 A0A2H1WC36 A0A2H1VZQ2 A0A194QNL9 A0A3S2PJD2 A0A2A4KBH0 A0A3Q0JEW1 A0A1B6DDS5 A0A232F572 A0A2A4KAH9 K7IXC8 A0A088AQ03 A0A195C9J1 A0A0C9RP37 A0A212FIH7 F4W9H3 Q26490 A0A194QL22 A0A224XS10 A0A3S2M7E4 E2AHR9 A0A067QQ48 A0A0N1IET4 A0A1B0Y0B2 A0A194QHU7 A0A0A9VT74 A0A069DSX1 E9GDV9 A0A2H1WC50 A0A0V0G967 A0A224XSY7 A0A212FIG6 A0A151IWM2 A0A0V0G6A3 K7IVH8 A0A195CPN9 A0A224XN75 A0A194QHR0 A0A224XRN0 A0A0M8ZSK1 A0A069DSW0 E2BKD5 A0A151WNL9 A0A151IPX0 H9JBS6 A0A0P5BW27 A0A0P5BGU9 A0A0P5BKL0 E2BKD3 A0A0P5BS40 A0A195DUR6 A0A0P5G8H7 A0A0P5Y1N1 A0A0P5Z523 A0A0P5C3I9 A0A0N8D9T5 A0A224XU45 A0A0P5LRK9 A0A164ZQS0 A0A0P5SY28 A0A0N8BUL6 A0A224XTD2 A0A0P5RQL9 A0A0P6FPG4 A0A0P5N4Y8 A0A3L8E1L6 A0A069DSB1 A0A0P5BUH5 A0A0P6EDE3 A0A023F783
PDB
1X8L
E-value=3.81201e-60,
Score=586
Ontologies
GO
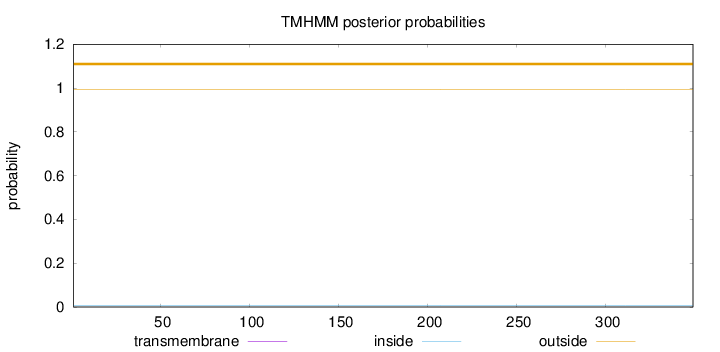
Topology
Length:
349
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00189
Exp number, first 60 AAs:
0.00011
Total prob of N-in:
0.00777
outside
1 - 349
Population Genetic Test Statistics
Pi
169.776679
Theta
149.987367
Tajima's D
0.207709
CLR
0.055464
CSRT
0.425878706064697
Interpretation
Uncertain
