Pre Gene Modal
BGIBMGA007138
Annotation
PREDICTED:_uncharacterized_protein_LOC101740749_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.043 PlasmaMembrane Reliability : 1.675
Sequence
CDS
ATGGAGGAATGTCGTGTGCAAAATAAATTGAAACTACTGTTTGAGAAACTTCTTGAACCGAATTATGTACTTAATAGTTATTATGCTGATGGCTTAATAAACAAACTGACGAATGCCAGCACGGATCCAGAAATCTCCTTTCTCGGATCATTACCGTTTTTAACTAATCTATTATTGGAAGCGATAAACAAAAGTGAAAATGCTCACGTCAGCGCCAAAGTATTTCTGACGAGAGTACTCGGGATCGTTTGCAAAACGGAACTCAATTTCACGAAGTTCAATTGTCCTCAAGGGGACATAATATTACAGGAACTTAAGCGTATCAACTCTCCCAATATCAATCCCAGTCTCAGGGTAGCATTCACGGAGGTAGCTCTGGCCATTGTCAACCACAGTTCAGGAGTTTCTTGGCTCTTAGAAACTGGGGTGTGGAAAGAAATCCTAAGTTTGGTCAATGAAAAAACAACGATCTTCGTGATAAGACAAGCGTATAAATTCGCTGCAGAATTCGTGTGGAAGCTAAATGATTTGAGAGACGTAAACAGTATCAAAGAAGTTATTAGTTATATAATAAGGCCAGTTAGCGAGGAAGATTACATGGGAATGAAAGTCTTGACAAGTGACGACGTAGAAGAAAAATGCAAGAGCTTAGAGCCAATGGCGAATATGTTAATAGCCATAATCAGCAAAGACAATCGTCTAGAACAACCCACCTTACTTCTAAGCATGATAGTCAAGGAGTTTAAGATTTCGCATTACTTGTATGTGGTAATAGACAGGATCCACCACGATGAAACGTCGTTGCTACTGTCCAAGGTTATCTTTTGGGTTGCAATAGTGAAGATTTTCCTGGAAAAACCCGCGAAACCCGGTGTCGAGTATGTCAGAGAGGATTTTCGCGATTTAGGGATCGTGTTCTTCAACGCTGTACAGTTTTTCATGTCGAGGCGCAACGCTTCGTGCGTAATCAGTTTCTCTGCGTTCTGTACCGTTATTTGGAAGATAGTTTTTGGTGATAAAACTGTCGAGAAATATGGCAAAGAACACGGAACGAGACAGATCAATTTGCGTAGCCAAAGTTTGTTTATGTGCATCGTTCCCATACTGGTGTTTATCAAGCTAGGGAAACCACCATCGGAGGCGACGCGCGAAAACATAAACGATTACATAGTGACATTATTGAATTCGTCGTGCGAATCCACGGCCAGGATATCGTTCGCGCTGAGAGATCTGATCTTAGAGTTGGACACGATGCCGCTTATCGTGCAGAGCGTGAAGAAGCTCACCTGTTTGAAACATCGCTTAAATGACGAGCAAGCGAACCTTGTGTTCCAGGCTTTATTTTACGCTCTGAGGGACTACGACCCTATAGACGACTTCGGCGACGTCAAACCGGATTATTCTTCGGTCGAAAATCAAGATAACGTCTTCGTGATGACCTACGTCCTCGACATCGTTCTGTTCCTCGTCAAGAATCACAATATAAACTGGCACGAAAGTATAGAAGTACTTTGTTTGTATTCCGTCGTCTATAATATTCTAAAGAAACCTAATTTAACTGTTAAGTTCGTAGTGACAGCCCTGAATGTAATAACCGTGACGATTAAGAAGTTCCTCCCGCCGAATCTGTCGCTGCTGCTGGAGTCGAAGCCGGGCTCCACGATGCACGAGCTCGGGAAACTGATCTACATGAAGCTACACGACCTACACTGGGAGGTGCGGGACTCCGCGCTCGAGTTGTTGCACGTTATTACAGAGATCTCGTTTATAAAATTCCCACCGTTCCAGAAGCAAATAATACAGGACAAACTCATAAACGTGGCCGCGACGGTCGCCTTCAACGACTACGAGGCTTACGTTCAAGTTTCTGCGCTCAAGTGTATCGGGGCTGCCAGCAAGGTCAACACTATATGGGAACTGTTGACCGCTGAGTTTCCTACAATTCAGGAACAAGCCGTCAGTATATTACGCAACAACCCGGAAGGAATCGTACGCAAAGAAGCTTGCAACGTATTGTGTGACTTGTACCAGAACTTAAAAATTAGTCCAAACTTCAAACAAGTGATATACGAACACATGGTGTCGTCTGCAATTTGCGATTTCCACTGGGAAGTCCAGTTGAGTGCTTTAAGGTTTTGGAAGATAGTTTTAGAATCATTGCTGACCGACCAAGGAATGTTGGACGGTGTATTCCCCCCAGTTACGTTTTCAAGGCAGTCGAGAAAAATAGTTACTCTGAATGAAACCGAAATTCAAAGACGATTGTTTCAAATACTAAACGAACTCAGTTCCAAAGGATGCTTGACTGTATTAGTAAAGTTGTTACATGACGATACAGAATCAGAGATTATGGAGAGTGCCTTAAATATATGTGATGAATTATTAGAAATTCTTAACAAACATAAAGTTCCAGAAAGTCTTAATGCAAGACCTGGTGACCCGCAATCGATAGACGAATTGCTATCGACGATTAAAGAAGAATCAGATAGCAATAGTAATTCGGAAGAAATGGCCGATGTCGACACTGCAACCGCTTCTGATAGTGTTATTGATGGGATACTAAAAGCAGACGACATAAACTTATTAGCAAACATATATGAGAGGCACATGTCACTGCAAACTGAAGTACAGATACCGGAGAAACAATTAAAACCAAAAATGAAACTGTTACAATTGGTAACTCCAAGTCTTTTTGTCAATTACCTGAAGAGTAAAGATTTTAAACAGGTTATTAAAGAGAAGAGGCGTTGGTCTGAAGGTATTAGAAGTATTTCATCATTACTTGATGATGTACTCGGCATGTATGATGTGGACGATGAAATCAATTCCTTGGACTGTTATTGA
Protein
MEECRVQNKLKLLFEKLLEPNYVLNSYYADGLINKLTNASTDPEISFLGSLPFLTNLLLEAINKSENAHVSAKVFLTRVLGIVCKTELNFTKFNCPQGDIILQELKRINSPNINPSLRVAFTEVALAIVNHSSGVSWLLETGVWKEILSLVNEKTTIFVIRQAYKFAAEFVWKLNDLRDVNSIKEVISYIIRPVSEEDYMGMKVLTSDDVEEKCKSLEPMANMLIAIISKDNRLEQPTLLLSMIVKEFKISHYLYVVIDRIHHDETSLLLSKVIFWVAIVKIFLEKPAKPGVEYVREDFRDLGIVFFNAVQFFMSRRNASCVISFSAFCTVIWKIVFGDKTVEKYGKEHGTRQINLRSQSLFMCIVPILVFIKLGKPPSEATRENINDYIVTLLNSSCESTARISFALRDLILELDTMPLIVQSVKKLTCLKHRLNDEQANLVFQALFYALRDYDPIDDFGDVKPDYSSVENQDNVFVMTYVLDIVLFLVKNHNINWHESIEVLCLYSVVYNILKKPNLTVKFVVTALNVITVTIKKFLPPNLSLLLESKPGSTMHELGKLIYMKLHDLHWEVRDSALELLHVITEISFIKFPPFQKQIIQDKLINVAATVAFNDYEAYVQVSALKCIGAASKVNTIWELLTAEFPTIQEQAVSILRNNPEGIVRKEACNVLCDLYQNLKISPNFKQVIYEHMVSSAICDFHWEVQLSALRFWKIVLESLLTDQGMLDGVFPPVTFSRQSRKIVTLNETEIQRRLFQILNELSSKGCLTVLVKLLHDDTESEIMESALNICDELLEILNKHKVPESLNARPGDPQSIDELLSTIKEESDSNSNSEEMADVDTATASDSVIDGILKADDINLLANIYERHMSLQTEVQIPEKQLKPKMKLLQLVTPSLFVNYLKSKDFKQVIKEKRRWSEGIRSISSLLDDVLGMYDVDDEINSLDCY
Summary
Uniprot
H9JC93
A0A2A4J3Y0
A0A2H1W8B9
A0A1E1W374
A0A1E1W3R1
A0A1E1W6I9
+ More
A0A1E1W3T1 A0A194RJC1 A0A194PIA0 A0A3S2PF00 A0A212EI32 A0A2A4J5M7 A0A2H1W8E3 A0A194PIJ3 A0A0K8TN05 A0A194RIK1 A0A1L8DDE8 W5JLP3 A0A1A9VIF0 A0A2M4AAU3 A0A2M4BBW1 A0A2M4BBT3 A0A182F920 A0A2M4BBU9 A0A1I8MQX0 A0A1Q3F5W9 A0A1B0B1V7 T1PGU0 A0A1A9ZPU6 B0WL50 A0A1A9W9D7 A0A1B0FFB0 A0A084WTX9 A0A182VKK2 A0A182J995 Q7PQ81 A0A182XIG2 A0A182KZG8 A0A182MFV2 A0A182PHW4 A0A182QT63 A0A182RE17 A0A2J7PK35 A0A182N9X3 Q16QC3 A0A182HYR6 A0A182UFN8 A0A182SFP7 A0A1J1HPS6 A0A182Y8Q3 A0A182H9N2 A0A0A1WED1 A0A195CWZ5 A0A0L0CK01 A0A195BRW7 A0A158NM13 F4WHT7 A0A182W3I5 A0A1Y1M8B4 A0A151JP09 A0A182K0F5 A0A1L8DBD5 B3P2Y5 B4PQA5 Q9VDE8 E2AZM1 B4NH52 A0A1I8NWR9 B3M2W4 W8BME1 B4R149 A0A151JVX3 A0A026WF00 A0A1W4VZU6 A0A154PIX2 B4KDM1 A0A088ARD3 E2B484 A0A0L7RF07 E9J4Y3 Q293K5 B4IKL2 A0A2A3EAJ8 A0A0K8W3B8 A0A0K8VMQ1 B4JFJ2 A0A3B0JR11 A0A3B0JSL3 A0A3B0JMA4 A0A0T6B792 A0A232F1Y9 B4LWV3 A0A0M4EKS9 B4GM02
A0A1E1W3T1 A0A194RJC1 A0A194PIA0 A0A3S2PF00 A0A212EI32 A0A2A4J5M7 A0A2H1W8E3 A0A194PIJ3 A0A0K8TN05 A0A194RIK1 A0A1L8DDE8 W5JLP3 A0A1A9VIF0 A0A2M4AAU3 A0A2M4BBW1 A0A2M4BBT3 A0A182F920 A0A2M4BBU9 A0A1I8MQX0 A0A1Q3F5W9 A0A1B0B1V7 T1PGU0 A0A1A9ZPU6 B0WL50 A0A1A9W9D7 A0A1B0FFB0 A0A084WTX9 A0A182VKK2 A0A182J995 Q7PQ81 A0A182XIG2 A0A182KZG8 A0A182MFV2 A0A182PHW4 A0A182QT63 A0A182RE17 A0A2J7PK35 A0A182N9X3 Q16QC3 A0A182HYR6 A0A182UFN8 A0A182SFP7 A0A1J1HPS6 A0A182Y8Q3 A0A182H9N2 A0A0A1WED1 A0A195CWZ5 A0A0L0CK01 A0A195BRW7 A0A158NM13 F4WHT7 A0A182W3I5 A0A1Y1M8B4 A0A151JP09 A0A182K0F5 A0A1L8DBD5 B3P2Y5 B4PQA5 Q9VDE8 E2AZM1 B4NH52 A0A1I8NWR9 B3M2W4 W8BME1 B4R149 A0A151JVX3 A0A026WF00 A0A1W4VZU6 A0A154PIX2 B4KDM1 A0A088ARD3 E2B484 A0A0L7RF07 E9J4Y3 Q293K5 B4IKL2 A0A2A3EAJ8 A0A0K8W3B8 A0A0K8VMQ1 B4JFJ2 A0A3B0JR11 A0A3B0JSL3 A0A3B0JMA4 A0A0T6B792 A0A232F1Y9 B4LWV3 A0A0M4EKS9 B4GM02
Pubmed
19121390
26354079
22118469
26369729
20920257
23761445
+ More
25315136 24438588 12364791 20966253 17510324 25244985 26483478 25830018 26108605 21347285 21719571 28004739 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20798317 24495485 24508170 30249741 21282665 15632085 28648823
25315136 24438588 12364791 20966253 17510324 25244985 26483478 25830018 26108605 21347285 21719571 28004739 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20798317 24495485 24508170 30249741 21282665 15632085 28648823
EMBL
BABH01015101
BABH01015102
NWSH01003243
PCG66701.1
ODYU01006991
SOQ49339.1
+ More
GDQN01009621 JAT81433.1 GDQN01009485 JAT81569.1 GDQN01008495 JAT82559.1 GDQN01009421 JAT81633.1 KQ460124 KPJ17652.1 KQ459603 KPI93136.1 RSAL01000062 RVE49536.1 AGBW02014713 OWR41138.1 PCG66702.1 SOQ49338.1 KPI93137.1 GDAI01002087 JAI15516.1 KPJ17653.1 GFDF01009724 JAV04360.1 ADMH02001067 ETN64223.1 GGFK01004576 MBW37897.1 GGFJ01001361 MBW50502.1 GGFJ01001364 MBW50505.1 GGFJ01001362 MBW50503.1 GFDL01012080 JAV22965.1 JXJN01007344 KA647971 AFP62600.1 DS231979 EDS30213.1 CCAG010008558 ATLV01026943 KE525420 KFB53673.1 AAAB01008898 EAA09193.6 AXCM01000353 AXCN02001202 NEVH01024942 PNF16693.1 CH477752 EAT36605.1 EAT36606.1 APCN01005402 CVRI01000006 CRK88217.1 JXUM01121251 JXUM01121252 JXUM01121253 JXUM01121254 KQ566479 KXJ70099.1 GBXI01017524 JAC96767.1 KQ977171 KYN05170.1 JRES01000301 KNC32575.1 KQ976421 KYM89107.1 ADTU01020139 GL888167 EGI66294.1 GEZM01037790 JAV81933.1 KQ978786 KYN28360.1 GFDF01010311 JAV03773.1 CH954181 EDV48299.1 CM000160 EDW96214.1 AE014297 BT044095 AAF55846.1 ACH92160.1 GL444254 EFN61120.1 CH964272 EDW84549.1 CH902617 EDV43494.1 GAMC01004290 JAC02266.1 CM000364 EDX12136.1 KQ981688 KYN37921.1 KK107238 QOIP01000007 EZA54607.1 RLU20121.1 KQ434931 KZC11785.1 CH933806 EDW13855.1 GL445515 EFN89501.1 KQ414608 KOC69413.1 GL768121 EFZ12122.1 CM000070 EAL29210.1 CH480853 EDW51616.1 KZ288308 PBC28738.1 GDHF01006777 JAI45537.1 GDHF01025845 GDHF01012147 JAI26469.1 JAI40167.1 CH916369 EDV93473.1 OUUW01000007 SPP83403.1 SPP83402.1 SPP83404.1 LJIG01009368 KRT83220.1 NNAY01001206 OXU24734.1 CH940650 EDW66674.1 CP012526 ALC45765.1 CH479185 EDW38576.1
GDQN01009621 JAT81433.1 GDQN01009485 JAT81569.1 GDQN01008495 JAT82559.1 GDQN01009421 JAT81633.1 KQ460124 KPJ17652.1 KQ459603 KPI93136.1 RSAL01000062 RVE49536.1 AGBW02014713 OWR41138.1 PCG66702.1 SOQ49338.1 KPI93137.1 GDAI01002087 JAI15516.1 KPJ17653.1 GFDF01009724 JAV04360.1 ADMH02001067 ETN64223.1 GGFK01004576 MBW37897.1 GGFJ01001361 MBW50502.1 GGFJ01001364 MBW50505.1 GGFJ01001362 MBW50503.1 GFDL01012080 JAV22965.1 JXJN01007344 KA647971 AFP62600.1 DS231979 EDS30213.1 CCAG010008558 ATLV01026943 KE525420 KFB53673.1 AAAB01008898 EAA09193.6 AXCM01000353 AXCN02001202 NEVH01024942 PNF16693.1 CH477752 EAT36605.1 EAT36606.1 APCN01005402 CVRI01000006 CRK88217.1 JXUM01121251 JXUM01121252 JXUM01121253 JXUM01121254 KQ566479 KXJ70099.1 GBXI01017524 JAC96767.1 KQ977171 KYN05170.1 JRES01000301 KNC32575.1 KQ976421 KYM89107.1 ADTU01020139 GL888167 EGI66294.1 GEZM01037790 JAV81933.1 KQ978786 KYN28360.1 GFDF01010311 JAV03773.1 CH954181 EDV48299.1 CM000160 EDW96214.1 AE014297 BT044095 AAF55846.1 ACH92160.1 GL444254 EFN61120.1 CH964272 EDW84549.1 CH902617 EDV43494.1 GAMC01004290 JAC02266.1 CM000364 EDX12136.1 KQ981688 KYN37921.1 KK107238 QOIP01000007 EZA54607.1 RLU20121.1 KQ434931 KZC11785.1 CH933806 EDW13855.1 GL445515 EFN89501.1 KQ414608 KOC69413.1 GL768121 EFZ12122.1 CM000070 EAL29210.1 CH480853 EDW51616.1 KZ288308 PBC28738.1 GDHF01006777 JAI45537.1 GDHF01025845 GDHF01012147 JAI26469.1 JAI40167.1 CH916369 EDV93473.1 OUUW01000007 SPP83403.1 SPP83402.1 SPP83404.1 LJIG01009368 KRT83220.1 NNAY01001206 OXU24734.1 CH940650 EDW66674.1 CP012526 ALC45765.1 CH479185 EDW38576.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000283053
UP000007151
+ More
UP000000673 UP000078200 UP000069272 UP000095301 UP000092460 UP000092445 UP000002320 UP000091820 UP000092444 UP000030765 UP000075903 UP000075880 UP000007062 UP000076407 UP000075882 UP000075883 UP000075885 UP000075886 UP000075900 UP000235965 UP000075884 UP000008820 UP000075840 UP000075902 UP000075901 UP000183832 UP000076408 UP000069940 UP000249989 UP000078542 UP000037069 UP000078540 UP000005205 UP000007755 UP000075920 UP000078492 UP000075881 UP000008711 UP000002282 UP000000803 UP000000311 UP000007798 UP000095300 UP000007801 UP000000304 UP000078541 UP000053097 UP000279307 UP000192221 UP000076502 UP000009192 UP000005203 UP000008237 UP000053825 UP000001819 UP000001292 UP000242457 UP000001070 UP000268350 UP000215335 UP000008792 UP000092553 UP000008744
UP000000673 UP000078200 UP000069272 UP000095301 UP000092460 UP000092445 UP000002320 UP000091820 UP000092444 UP000030765 UP000075903 UP000075880 UP000007062 UP000076407 UP000075882 UP000075883 UP000075885 UP000075886 UP000075900 UP000235965 UP000075884 UP000008820 UP000075840 UP000075902 UP000075901 UP000183832 UP000076408 UP000069940 UP000249989 UP000078542 UP000037069 UP000078540 UP000005205 UP000007755 UP000075920 UP000078492 UP000075881 UP000008711 UP000002282 UP000000803 UP000000311 UP000007798 UP000095300 UP000007801 UP000000304 UP000078541 UP000053097 UP000279307 UP000192221 UP000076502 UP000009192 UP000005203 UP000008237 UP000053825 UP000001819 UP000001292 UP000242457 UP000001070 UP000268350 UP000215335 UP000008792 UP000092553 UP000008744
Interpro
SUPFAM
SSF48371
SSF48371
Gene 3D
ProteinModelPortal
H9JC93
A0A2A4J3Y0
A0A2H1W8B9
A0A1E1W374
A0A1E1W3R1
A0A1E1W6I9
+ More
A0A1E1W3T1 A0A194RJC1 A0A194PIA0 A0A3S2PF00 A0A212EI32 A0A2A4J5M7 A0A2H1W8E3 A0A194PIJ3 A0A0K8TN05 A0A194RIK1 A0A1L8DDE8 W5JLP3 A0A1A9VIF0 A0A2M4AAU3 A0A2M4BBW1 A0A2M4BBT3 A0A182F920 A0A2M4BBU9 A0A1I8MQX0 A0A1Q3F5W9 A0A1B0B1V7 T1PGU0 A0A1A9ZPU6 B0WL50 A0A1A9W9D7 A0A1B0FFB0 A0A084WTX9 A0A182VKK2 A0A182J995 Q7PQ81 A0A182XIG2 A0A182KZG8 A0A182MFV2 A0A182PHW4 A0A182QT63 A0A182RE17 A0A2J7PK35 A0A182N9X3 Q16QC3 A0A182HYR6 A0A182UFN8 A0A182SFP7 A0A1J1HPS6 A0A182Y8Q3 A0A182H9N2 A0A0A1WED1 A0A195CWZ5 A0A0L0CK01 A0A195BRW7 A0A158NM13 F4WHT7 A0A182W3I5 A0A1Y1M8B4 A0A151JP09 A0A182K0F5 A0A1L8DBD5 B3P2Y5 B4PQA5 Q9VDE8 E2AZM1 B4NH52 A0A1I8NWR9 B3M2W4 W8BME1 B4R149 A0A151JVX3 A0A026WF00 A0A1W4VZU6 A0A154PIX2 B4KDM1 A0A088ARD3 E2B484 A0A0L7RF07 E9J4Y3 Q293K5 B4IKL2 A0A2A3EAJ8 A0A0K8W3B8 A0A0K8VMQ1 B4JFJ2 A0A3B0JR11 A0A3B0JSL3 A0A3B0JMA4 A0A0T6B792 A0A232F1Y9 B4LWV3 A0A0M4EKS9 B4GM02
A0A1E1W3T1 A0A194RJC1 A0A194PIA0 A0A3S2PF00 A0A212EI32 A0A2A4J5M7 A0A2H1W8E3 A0A194PIJ3 A0A0K8TN05 A0A194RIK1 A0A1L8DDE8 W5JLP3 A0A1A9VIF0 A0A2M4AAU3 A0A2M4BBW1 A0A2M4BBT3 A0A182F920 A0A2M4BBU9 A0A1I8MQX0 A0A1Q3F5W9 A0A1B0B1V7 T1PGU0 A0A1A9ZPU6 B0WL50 A0A1A9W9D7 A0A1B0FFB0 A0A084WTX9 A0A182VKK2 A0A182J995 Q7PQ81 A0A182XIG2 A0A182KZG8 A0A182MFV2 A0A182PHW4 A0A182QT63 A0A182RE17 A0A2J7PK35 A0A182N9X3 Q16QC3 A0A182HYR6 A0A182UFN8 A0A182SFP7 A0A1J1HPS6 A0A182Y8Q3 A0A182H9N2 A0A0A1WED1 A0A195CWZ5 A0A0L0CK01 A0A195BRW7 A0A158NM13 F4WHT7 A0A182W3I5 A0A1Y1M8B4 A0A151JP09 A0A182K0F5 A0A1L8DBD5 B3P2Y5 B4PQA5 Q9VDE8 E2AZM1 B4NH52 A0A1I8NWR9 B3M2W4 W8BME1 B4R149 A0A151JVX3 A0A026WF00 A0A1W4VZU6 A0A154PIX2 B4KDM1 A0A088ARD3 E2B484 A0A0L7RF07 E9J4Y3 Q293K5 B4IKL2 A0A2A3EAJ8 A0A0K8W3B8 A0A0K8VMQ1 B4JFJ2 A0A3B0JR11 A0A3B0JSL3 A0A3B0JMA4 A0A0T6B792 A0A232F1Y9 B4LWV3 A0A0M4EKS9 B4GM02
Ontologies
GO
PANTHER
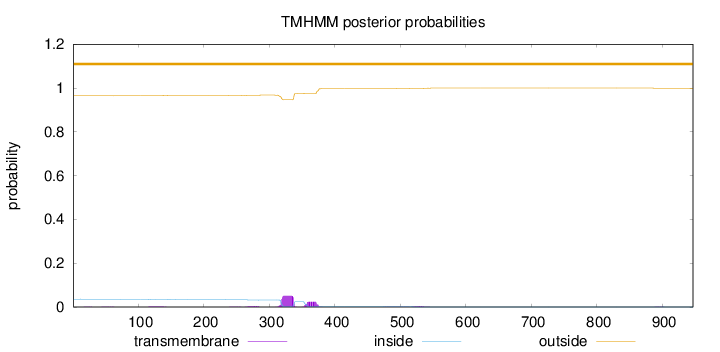
Topology
Length:
947
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.60601
Exp number, first 60 AAs:
0.01249
Total prob of N-in:
0.03574
outside
1 - 947
Population Genetic Test Statistics
Pi
55.814545
Theta
152.62991
Tajima's D
-1.337667
CLR
1535.752482
CSRT
0.080245987700615
Interpretation
Uncertain
