Gene
KWMTBOMO10543
Pre Gene Modal
BGIBMGA007137
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_vacuolar_protein_sorting-associated_protein_18_homolog_[Bombyx_mori]
Full name
Vacuolar protein sorting-associated protein 18 homolog
Location in the cell
Cytoplasmic Reliability : 1.75 Nuclear Reliability : 2.081
Sequence
CDS
ATGACTTCGATATTAGATCAGTATAAGCAAGCAGCACAGGCTTCCTATAGAACAAAGCCACCTGCTGAGCCCATGATATCTTCAGGATACATCAATATGCAGCTCGAAGATAATGTACCGATGTTTACGAAACAGAAAATGAACCTCAACCCATCGGATTTAATCACTCACGCTGCCGTTTCTAGTGATAACTTAGTGGTGGCTATGGCGAATGGGAAATTGTTTAGAATGGACATAAGGAATCCTGATTCTGAGCAAGAAATTCACTACTCAAAATATTGTCAGCCAACCTCTAAATTGAGCGGTTTGTTTCTGGATCCACTCGGATGTCATCTCTTAATGTCATTTTCATCTAAAACAAAAGATGGATGCCCTGAACTAGCCTATATCCATCAAAAGAGCTCGAAACTAAAATTTGTCAGCAAAATCCCCAACTATGAAATTACTGAAGTTGGTTGGAATTTTGAAAACACATCTAACAACATGACGGGACCTATTTTATTAGGCACTTCTAAAGGACATTTACTAGAAACTGAGTTAGAACCGAACAATGACAAAATGTTCATTGCCAGTGAACAGTACTGGCGACAGATTTTCGACATTGGAAAGGGGGCAGACACACCCATAACTGGAATAAAATTTCACAGGGTTGCCGGAACAAAAAAGTACTGTATATTTGTGACAACACCAACAAGAATTTACCAGTTCATCGGGCATGCCATTTTTTCTGAGAACAAACCATCTCTGAAGTCAGTCTTTTATCCGTATTTAACCATTCCAGAAACAGGTTTTCAGGAAATACCTTCTACGTTAAAATATTCTGAGCTACAATATTACTTTGATAAGAATGAAATGCCTAAAACGTTTGCCTGGCTTACCGAACAGGGTATCTTCTATGGTCAGTTGGACCCGACATCTCAACAGAACTCAAATTCTGTTGTCACCCAAGGCTCGCTCGTAACCTACCCTTCGGGACCGGACGGCAAAGAAATACCCCCATTATCATTTGTTCTCACGGAATTCCACACTCTGCTAATGTATTCAGATCGAGTTAAAGTAGTATCTCTACTCAATCAAGAACTTGTCCACGAGGATATTTACTCGGAAGCTCATGGGAAACTCAAAACAATAATACGCGACTTGAAGAAAAGACAGATTTGGGTGTTTACGGAAAAAAAGATATTTCGCTACAAGATCGTTCGTGAAGAACGGAACCTATGGCAGATATATTCAGATAAAGAACAGTTCGATCTGGCTAGAGAATATTGTCAGAACAATCCGGCCTTCCTGGATATCGTGAACATGAAACAAGCCGATCTGTTCTTCTCGAAAGGCGAATACGAGAGAAGTGCCGAAATATATGCCGAAACCCAGAGAAGCTTTGAGACGATATGTCTTAAGTTTTTGGAAGCCGAGCAATTTGATGCCTTAAAGAAATACTTGGAAAGGCGACTGGAGTCGTTGACAGATGACGATAAGACGTTGATCTCGATGATCGTTATCTGGATGACGGAGTTGTATCTGTCGCAACTGGGCACGCTGCGGCGGACGGGCAGGGGCGACAGCTCGGAATACCATCAGATGCAGAGCGACTTCGAAGTGTTCCTGCTCCATCCGAAGGTATCGAAGACGATGCGTCACGTTAAATCCGTCATATACGAGCTGATGTCCTCGCACGGCGACAAACAGAATCTACTGAAACTGACGATGATCAACGAGGATTTTGAAAACGTCGTGTCCCAGTACGTGTACAGGAAGAATTATCTAGAAGCTATAAACGCGCTGCAGAACTTGAAGCGCCCGGACCTGTTCTACCAGTTCGCTCCGGTGCTGATGGAGGCGGCTCCCAAGCACATGGTGAACGCTCTGATCGCTCAAGGCGCCAGGCTGAGTCCCTCGAAGCTGTTGCCGGCGTTCCTGTCCTGTCAGAACGACGAGGCGCACATCACGGAGATCACCAGATACTTGGAGTTCATTGTGGTGCACTTCAACGTGAAGGACAAGGCGATTCACAACTACTTGCTGACGCTGTACGCGGAGCATGATCAGCAGGCGCTGATGAGGTACCTGTCGCGGCAAGGGCAGGACATCGCGCTGGTCAACTACGATGTTCATTACGCGCTGAGGCTTTGTCGCGAGAAGAACGCGTCTGAGGCGTGCGTGAAGCTGTCGGCGCTGCTGGGGCTGTGGGAAGGCGCCGTGGAGCTGGCGCTGCGCGTGCGGCTCGCGCTCGCCAAGAGCGTCGCCGCGCTGCCCGACTGCCCGCACGTGCAGCGCCAGCTGTGGCTGCACATCGCTGAACACGTGATCACCAAGAACCAAGACATCAAGGAAGCTATGAGTTTCCTCGAGGAGTGTCCTCTGATCAAGATCGAAGATATCCTGCCGTTCTTCAGCGACGTGGTCACCATAGATCATTTTAGGGAACCTATTTGTCAATCTTTACAGGAGTACAACAATCAAATTGAGGAGACAAAAGCTGAAATGGAAGAAGCTACGAAGGCCGCTAAATACGTTCGCGATGAGATCCACTCGTTCCGCGGTCGCAGTGTGCGCGTGTCGGCCTCGTCCGGGTGCTGCGCCTGCGACCTCGCGCTGCTGCTGCGGCCCTTCTACCTCTTCCCCTGCGGACACCACTTCCACGCGGACTGCCTCACTGCCGAGATCACGCCGATGCTTGGTGCAGCAAAGCGCAGCAAGCTGGGCGACCTGCAGCGGCAGCTGTCCGTGCTGTCCAACATCGAGCTGTCCACCGCCACGTCGAGCGGCCTGCCCCTCAGGGAAGTCTTGAAGAACGAAATAGACGACATCGTGGCGAACGAATGCATCTACTGCGGAGAGCACATGATCAGTTGCATCGATAAACCGTTCATATCAGACGAGGACTGGGACCGAGTCATGAAGGAATGGGAGTAG
Protein
MTSILDQYKQAAQASYRTKPPAEPMISSGYINMQLEDNVPMFTKQKMNLNPSDLITHAAVSSDNLVVAMANGKLFRMDIRNPDSEQEIHYSKYCQPTSKLSGLFLDPLGCHLLMSFSSKTKDGCPELAYIHQKSSKLKFVSKIPNYEITEVGWNFENTSNNMTGPILLGTSKGHLLETELEPNNDKMFIASEQYWRQIFDIGKGADTPITGIKFHRVAGTKKYCIFVTTPTRIYQFIGHAIFSENKPSLKSVFYPYLTIPETGFQEIPSTLKYSELQYYFDKNEMPKTFAWLTEQGIFYGQLDPTSQQNSNSVVTQGSLVTYPSGPDGKEIPPLSFVLTEFHTLLMYSDRVKVVSLLNQELVHEDIYSEAHGKLKTIIRDLKKRQIWVFTEKKIFRYKIVREERNLWQIYSDKEQFDLAREYCQNNPAFLDIVNMKQADLFFSKGEYERSAEIYAETQRSFETICLKFLEAEQFDALKKYLERRLESLTDDDKTLISMIVIWMTELYLSQLGTLRRTGRGDSSEYHQMQSDFEVFLLHPKVSKTMRHVKSVIYELMSSHGDKQNLLKLTMINEDFENVVSQYVYRKNYLEAINALQNLKRPDLFYQFAPVLMEAAPKHMVNALIAQGARLSPSKLLPAFLSCQNDEAHITEITRYLEFIVVHFNVKDKAIHNYLLTLYAEHDQQALMRYLSRQGQDIALVNYDVHYALRLCREKNASEACVKLSALLGLWEGAVELALRVRLALAKSVAALPDCPHVQRQLWLHIAEHVITKNQDIKEAMSFLEECPLIKIEDILPFFSDVVTIDHFREPICQSLQEYNNQIEETKAEMEEATKAAKYVRDEIHSFRGRSVRVSASSGCCACDLALLLRPFYLFPCGHHFHADCLTAEITPMLGAAKRSKLGDLQRQLSVLSNIELSTATSSGLPLREVLKNEIDDIVANECIYCGEHMISCIDKPFISDEDWDRVMKEWE
Summary
Description
Plays a role in vesicle-mediated protein trafficking to lysosomal compartments including the endocytic membrane transport pathways. Believed to act as a core component of the putative HOPS endosomal tethering complex which is proposed to be involved in the Rab5-to-Rab7 endosome conversion probably implicating MON1A/B, and via binding SNAREs and SNARE complexes to mediate tethering and docking events during SNARE-mediated membrane fusion (By similarity). May be involved in vesicle trafficking to the hepatocyte apical membrane and play a role in development of the intra-hepatic biliary tree. May target endosomes to the pigment granule in melanocytes. Essential for early embryonic development.
Subunit
Component of the homotypic fusion and vacuole protein sorting (HOPS) complex (By similarity).
Similarity
Belongs to the VPS18 family.
Keywords
Coiled coil
Complete proteome
Developmental protein
Endosome
Lysosome
Membrane
Metal-binding
Protein transport
Reference proteome
Transport
Zinc
Zinc-finger
Feature
chain Vacuolar protein sorting-associated protein 18 homolog
Uniprot
H9JC92
A0A2A4JRZ0
A0A1E1WC00
A0A3S2NF81
A0A194RJY8
A0A194PK78
+ More
A0A2H1WDX1 A0A0L7LGG3 A0A212EI51 A0A2J7PK09 A0A2J7PK30 A0A2P8Y8K0 A0A026WFF8 F4WMW6 A0A3L8DKC3 A0A1W4WF15 A0A151I3N5 A0A2A3EMH9 A0A1B6FEP5 A0A067RGK1 A0A1W4W4G7 E2BRY6 E2ASS5 D6WXT3 A0A1W4WG09 A0A232F9I8 A0A0C9RYL8 K7J785 A0A151WIE3 A0A151IVV1 A0A1B6CTG6 A0A195FUI8 A0A088AC85 A0A1B6D9S6 A0A0L7RF67 A0A1Y1NJW9 N6TK23 U4UUW1 A0A158NXY7 E0VT90 A0A2R2MJ62 T1HVK6 A0A2L2YFR6 V4C6J3 R7V2G8 A0A1Q3G3T1 A0A210PZ54 A0A1S4F7C1 Q17DE2 A0A131YLT4 A0A182GQJ8 A0A224Z5C1 A0A182HAS5 L7M795 A0A1E1XC32 A0A1B6HNR0 A0A2C9LXL4 A0A1B0CQ76 A0A1Y1NPC1 A0A3R7PJJ1 A0A2S2QLW8 J9K4C7 S4RHG0 C3XXM3 A0A2H8TNL0 A8WGL1 B7ZVV5 P59015 H2U883 A0A293MVN3 A0A3Q3B983 A0A1B6DPR2 A0A2S2PM43 A0A2I4BCD6 A0A3B4U5E1 H3D060 A0A3P8WZV4 A0A087Y4U4 W5U6C4 A0A3Q2QYR8 A0A146ZN76 A0A3Q1GBY4 A0A3Q2YIG6 A0A3B3R5R9 I3IWH5 A0A315VX64 A0A146MP47 A0A3P8NY87 A0A3P9C6D1 A0A3B3XTV8 A0A3Q1B4F1 A0A3P8U4F8 A0A131XZF8 A0A147BHD1 M3ZX52 A0A3Q3MCT9 A0A3B5LHS2 W5LD51 A0A1A7Z3T4 A0A3P9PDV4
A0A2H1WDX1 A0A0L7LGG3 A0A212EI51 A0A2J7PK09 A0A2J7PK30 A0A2P8Y8K0 A0A026WFF8 F4WMW6 A0A3L8DKC3 A0A1W4WF15 A0A151I3N5 A0A2A3EMH9 A0A1B6FEP5 A0A067RGK1 A0A1W4W4G7 E2BRY6 E2ASS5 D6WXT3 A0A1W4WG09 A0A232F9I8 A0A0C9RYL8 K7J785 A0A151WIE3 A0A151IVV1 A0A1B6CTG6 A0A195FUI8 A0A088AC85 A0A1B6D9S6 A0A0L7RF67 A0A1Y1NJW9 N6TK23 U4UUW1 A0A158NXY7 E0VT90 A0A2R2MJ62 T1HVK6 A0A2L2YFR6 V4C6J3 R7V2G8 A0A1Q3G3T1 A0A210PZ54 A0A1S4F7C1 Q17DE2 A0A131YLT4 A0A182GQJ8 A0A224Z5C1 A0A182HAS5 L7M795 A0A1E1XC32 A0A1B6HNR0 A0A2C9LXL4 A0A1B0CQ76 A0A1Y1NPC1 A0A3R7PJJ1 A0A2S2QLW8 J9K4C7 S4RHG0 C3XXM3 A0A2H8TNL0 A8WGL1 B7ZVV5 P59015 H2U883 A0A293MVN3 A0A3Q3B983 A0A1B6DPR2 A0A2S2PM43 A0A2I4BCD6 A0A3B4U5E1 H3D060 A0A3P8WZV4 A0A087Y4U4 W5U6C4 A0A3Q2QYR8 A0A146ZN76 A0A3Q1GBY4 A0A3Q2YIG6 A0A3B3R5R9 I3IWH5 A0A315VX64 A0A146MP47 A0A3P8NY87 A0A3P9C6D1 A0A3B3XTV8 A0A3Q1B4F1 A0A3P8U4F8 A0A131XZF8 A0A147BHD1 M3ZX52 A0A3Q3MCT9 A0A3B5LHS2 W5LD51 A0A1A7Z3T4 A0A3P9PDV4
Pubmed
19121390
26354079
26227816
22118469
29403074
24508170
+ More
21719571 30249741 24845553 20798317 18362917 19820115 28648823 20075255 28004739 23537049 21347285 20566863 26561354 23254933 28812685 17510324 26830274 26483478 28797301 25576852 28503490 15562597 18563158 23594743 12006978 16000385 21551351 15496914 24487278 23127152 29240929 25186727 29703783 29652888 23542700 25329095
21719571 30249741 24845553 20798317 18362917 19820115 28648823 20075255 28004739 23537049 21347285 20566863 26561354 23254933 28812685 17510324 26830274 26483478 28797301 25576852 28503490 15562597 18563158 23594743 12006978 16000385 21551351 15496914 24487278 23127152 29240929 25186727 29703783 29652888 23542700 25329095
EMBL
BABH01015094
NWSH01000689
PCG74771.1
GDQN01006551
JAT84503.1
RSAL01000062
+ More
RVE49538.1 KQ460124 KPJ17650.1 KQ459603 KPI93134.1 ODYU01007953 SOQ51166.1 JTDY01001179 KOB74633.1 AGBW02014713 OWR41140.1 NEVH01024942 PNF16670.1 PNF16669.1 PYGN01000810 PSN40464.1 KK107238 EZA54658.1 GL888223 EGI64436.1 QOIP01000007 RLU20633.1 KQ976482 KYM83709.1 KZ288210 PBC32965.1 GECZ01021109 JAS48660.1 KK852652 KDR19383.1 GL450100 EFN81545.1 GL442359 EFN63500.1 KQ971362 EFA08901.1 NNAY01000636 OXU27272.1 GBYB01013061 GBYB01013062 GBYB01013063 GBYB01013066 JAG82828.1 JAG82829.1 JAG82830.1 JAG82833.1 KQ983089 KYQ47639.1 KQ980882 KYN11920.1 GEDC01020590 JAS16708.1 KQ981261 KYN44118.1 GEDC01014854 JAS22444.1 KQ414608 KOC69473.1 GEZM01001278 JAV97999.1 APGK01021726 APGK01021727 KB740356 ENN80819.1 KB632375 ERL93965.1 ADTU01003708 ADTU01003709 ADTU01003710 DS235760 EEB16596.1 ACPB03014176 IAAA01010689 LAA06210.1 KB201305 ESO97284.1 AMQN01005238 KB295515 ELU13053.1 GFDL01000583 JAV34462.1 NEDP02005356 OWF41777.1 CH477296 EAT44395.1 GEDV01009686 JAP78871.1 JXUM01080879 JXUM01080880 KQ563214 KXJ74279.1 GFPF01013259 MAA24405.1 JXUM01031405 JXUM01031406 KQ560920 KXJ80346.1 GACK01005297 JAA59737.1 GFAC01002386 JAT96802.1 GECU01031471 JAS76235.1 AJWK01023187 AJWK01023188 GEZM01001279 JAV97997.1 QCYY01002652 ROT68617.1 GGMS01009544 MBY78747.1 ABLF02029049 GG666471 EEN67483.1 GFXV01003417 MBW15222.1 BC154756 AAI54757.1 BC171720 AAI71720.1 AL929049 BX897725 AF506204 BC086828 GFWV01019270 MAA43998.1 GEDC01009636 JAS27662.1 GGMR01017828 MBY30447.1 AYCK01008713 AYCK01008714 JT405787 AHH37325.1 GCES01018534 JAR67789.1 AERX01005499 NHOQ01001000 PWA27813.1 GCES01164898 JAQ21424.1 GEFM01004164 JAP71632.1 GEGO01005211 JAR90193.1 HADW01004454 HADX01014784 SBP37016.1
RVE49538.1 KQ460124 KPJ17650.1 KQ459603 KPI93134.1 ODYU01007953 SOQ51166.1 JTDY01001179 KOB74633.1 AGBW02014713 OWR41140.1 NEVH01024942 PNF16670.1 PNF16669.1 PYGN01000810 PSN40464.1 KK107238 EZA54658.1 GL888223 EGI64436.1 QOIP01000007 RLU20633.1 KQ976482 KYM83709.1 KZ288210 PBC32965.1 GECZ01021109 JAS48660.1 KK852652 KDR19383.1 GL450100 EFN81545.1 GL442359 EFN63500.1 KQ971362 EFA08901.1 NNAY01000636 OXU27272.1 GBYB01013061 GBYB01013062 GBYB01013063 GBYB01013066 JAG82828.1 JAG82829.1 JAG82830.1 JAG82833.1 KQ983089 KYQ47639.1 KQ980882 KYN11920.1 GEDC01020590 JAS16708.1 KQ981261 KYN44118.1 GEDC01014854 JAS22444.1 KQ414608 KOC69473.1 GEZM01001278 JAV97999.1 APGK01021726 APGK01021727 KB740356 ENN80819.1 KB632375 ERL93965.1 ADTU01003708 ADTU01003709 ADTU01003710 DS235760 EEB16596.1 ACPB03014176 IAAA01010689 LAA06210.1 KB201305 ESO97284.1 AMQN01005238 KB295515 ELU13053.1 GFDL01000583 JAV34462.1 NEDP02005356 OWF41777.1 CH477296 EAT44395.1 GEDV01009686 JAP78871.1 JXUM01080879 JXUM01080880 KQ563214 KXJ74279.1 GFPF01013259 MAA24405.1 JXUM01031405 JXUM01031406 KQ560920 KXJ80346.1 GACK01005297 JAA59737.1 GFAC01002386 JAT96802.1 GECU01031471 JAS76235.1 AJWK01023187 AJWK01023188 GEZM01001279 JAV97997.1 QCYY01002652 ROT68617.1 GGMS01009544 MBY78747.1 ABLF02029049 GG666471 EEN67483.1 GFXV01003417 MBW15222.1 BC154756 AAI54757.1 BC171720 AAI71720.1 AL929049 BX897725 AF506204 BC086828 GFWV01019270 MAA43998.1 GEDC01009636 JAS27662.1 GGMR01017828 MBY30447.1 AYCK01008713 AYCK01008714 JT405787 AHH37325.1 GCES01018534 JAR67789.1 AERX01005499 NHOQ01001000 PWA27813.1 GCES01164898 JAQ21424.1 GEFM01004164 JAP71632.1 GEGO01005211 JAR90193.1 HADW01004454 HADX01014784 SBP37016.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000053268
UP000037510
+ More
UP000007151 UP000235965 UP000245037 UP000053097 UP000007755 UP000279307 UP000192223 UP000078540 UP000242457 UP000027135 UP000008237 UP000000311 UP000007266 UP000215335 UP000002358 UP000075809 UP000078492 UP000078541 UP000005203 UP000053825 UP000019118 UP000030742 UP000005205 UP000009046 UP000085678 UP000015103 UP000030746 UP000014760 UP000242188 UP000008820 UP000069940 UP000249989 UP000076420 UP000092461 UP000283509 UP000007819 UP000245300 UP000001554 UP000000437 UP000005226 UP000264800 UP000192220 UP000261420 UP000007303 UP000265120 UP000028760 UP000221080 UP000265000 UP000257200 UP000264820 UP000261540 UP000005207 UP000265100 UP000265160 UP000261480 UP000257160 UP000265080 UP000002852 UP000261640 UP000261380 UP000018467 UP000242638
UP000007151 UP000235965 UP000245037 UP000053097 UP000007755 UP000279307 UP000192223 UP000078540 UP000242457 UP000027135 UP000008237 UP000000311 UP000007266 UP000215335 UP000002358 UP000075809 UP000078492 UP000078541 UP000005203 UP000053825 UP000019118 UP000030742 UP000005205 UP000009046 UP000085678 UP000015103 UP000030746 UP000014760 UP000242188 UP000008820 UP000069940 UP000249989 UP000076420 UP000092461 UP000283509 UP000007819 UP000245300 UP000001554 UP000000437 UP000005226 UP000264800 UP000192220 UP000261420 UP000007303 UP000265120 UP000028760 UP000221080 UP000265000 UP000257200 UP000264820 UP000261540 UP000005207 UP000265100 UP000265160 UP000261480 UP000257160 UP000265080 UP000002852 UP000261640 UP000261380 UP000018467 UP000242638
Interpro
IPR016024
ARM-type_fold
+ More
IPR000547 Clathrin_H-chain/VPS_repeat
IPR007810 Pep3_Vps18
IPR013083 Znf_RING/FYVE/PHD
IPR011990 TPR-like_helical_dom_sf
IPR036322 WD40_repeat_dom_sf
IPR002562 3'-5'_exonuclease_dom
IPR012337 RNaseH-like_sf
IPR036397 RNaseH_sf
IPR019453 VPS39/TGF_beta_rcpt-assoc_2
IPR001841 Znf_RING
IPR000547 Clathrin_H-chain/VPS_repeat
IPR007810 Pep3_Vps18
IPR013083 Znf_RING/FYVE/PHD
IPR011990 TPR-like_helical_dom_sf
IPR036322 WD40_repeat_dom_sf
IPR002562 3'-5'_exonuclease_dom
IPR012337 RNaseH-like_sf
IPR036397 RNaseH_sf
IPR019453 VPS39/TGF_beta_rcpt-assoc_2
IPR001841 Znf_RING
Gene 3D
ProteinModelPortal
H9JC92
A0A2A4JRZ0
A0A1E1WC00
A0A3S2NF81
A0A194RJY8
A0A194PK78
+ More
A0A2H1WDX1 A0A0L7LGG3 A0A212EI51 A0A2J7PK09 A0A2J7PK30 A0A2P8Y8K0 A0A026WFF8 F4WMW6 A0A3L8DKC3 A0A1W4WF15 A0A151I3N5 A0A2A3EMH9 A0A1B6FEP5 A0A067RGK1 A0A1W4W4G7 E2BRY6 E2ASS5 D6WXT3 A0A1W4WG09 A0A232F9I8 A0A0C9RYL8 K7J785 A0A151WIE3 A0A151IVV1 A0A1B6CTG6 A0A195FUI8 A0A088AC85 A0A1B6D9S6 A0A0L7RF67 A0A1Y1NJW9 N6TK23 U4UUW1 A0A158NXY7 E0VT90 A0A2R2MJ62 T1HVK6 A0A2L2YFR6 V4C6J3 R7V2G8 A0A1Q3G3T1 A0A210PZ54 A0A1S4F7C1 Q17DE2 A0A131YLT4 A0A182GQJ8 A0A224Z5C1 A0A182HAS5 L7M795 A0A1E1XC32 A0A1B6HNR0 A0A2C9LXL4 A0A1B0CQ76 A0A1Y1NPC1 A0A3R7PJJ1 A0A2S2QLW8 J9K4C7 S4RHG0 C3XXM3 A0A2H8TNL0 A8WGL1 B7ZVV5 P59015 H2U883 A0A293MVN3 A0A3Q3B983 A0A1B6DPR2 A0A2S2PM43 A0A2I4BCD6 A0A3B4U5E1 H3D060 A0A3P8WZV4 A0A087Y4U4 W5U6C4 A0A3Q2QYR8 A0A146ZN76 A0A3Q1GBY4 A0A3Q2YIG6 A0A3B3R5R9 I3IWH5 A0A315VX64 A0A146MP47 A0A3P8NY87 A0A3P9C6D1 A0A3B3XTV8 A0A3Q1B4F1 A0A3P8U4F8 A0A131XZF8 A0A147BHD1 M3ZX52 A0A3Q3MCT9 A0A3B5LHS2 W5LD51 A0A1A7Z3T4 A0A3P9PDV4
A0A2H1WDX1 A0A0L7LGG3 A0A212EI51 A0A2J7PK09 A0A2J7PK30 A0A2P8Y8K0 A0A026WFF8 F4WMW6 A0A3L8DKC3 A0A1W4WF15 A0A151I3N5 A0A2A3EMH9 A0A1B6FEP5 A0A067RGK1 A0A1W4W4G7 E2BRY6 E2ASS5 D6WXT3 A0A1W4WG09 A0A232F9I8 A0A0C9RYL8 K7J785 A0A151WIE3 A0A151IVV1 A0A1B6CTG6 A0A195FUI8 A0A088AC85 A0A1B6D9S6 A0A0L7RF67 A0A1Y1NJW9 N6TK23 U4UUW1 A0A158NXY7 E0VT90 A0A2R2MJ62 T1HVK6 A0A2L2YFR6 V4C6J3 R7V2G8 A0A1Q3G3T1 A0A210PZ54 A0A1S4F7C1 Q17DE2 A0A131YLT4 A0A182GQJ8 A0A224Z5C1 A0A182HAS5 L7M795 A0A1E1XC32 A0A1B6HNR0 A0A2C9LXL4 A0A1B0CQ76 A0A1Y1NPC1 A0A3R7PJJ1 A0A2S2QLW8 J9K4C7 S4RHG0 C3XXM3 A0A2H8TNL0 A8WGL1 B7ZVV5 P59015 H2U883 A0A293MVN3 A0A3Q3B983 A0A1B6DPR2 A0A2S2PM43 A0A2I4BCD6 A0A3B4U5E1 H3D060 A0A3P8WZV4 A0A087Y4U4 W5U6C4 A0A3Q2QYR8 A0A146ZN76 A0A3Q1GBY4 A0A3Q2YIG6 A0A3B3R5R9 I3IWH5 A0A315VX64 A0A146MP47 A0A3P8NY87 A0A3P9C6D1 A0A3B3XTV8 A0A3Q1B4F1 A0A3P8U4F8 A0A131XZF8 A0A147BHD1 M3ZX52 A0A3Q3MCT9 A0A3B5LHS2 W5LD51 A0A1A7Z3T4 A0A3P9PDV4
PDB
4UUY
E-value=5.54859e-07,
Score=132
Ontologies
GO
GO:0006886
GO:0016192
GO:0005623
GO:0005768
GO:0035542
GO:0030897
GO:0007033
GO:0007032
GO:0006904
GO:0007040
GO:0030674
GO:0008408
GO:0003676
GO:0016021
GO:0043485
GO:0015721
GO:0048069
GO:0060036
GO:0007634
GO:0045176
GO:0008333
GO:0031902
GO:0005765
GO:0046872
GO:0048757
GO:0008152
GO:0050660
GO:0006284
GO:0003906
GO:0005634
GO:0006285
GO:0019104
GO:0003824
Topology
Subcellular location
Late endosome membrane
Lysosome membrane
Lysosome membrane
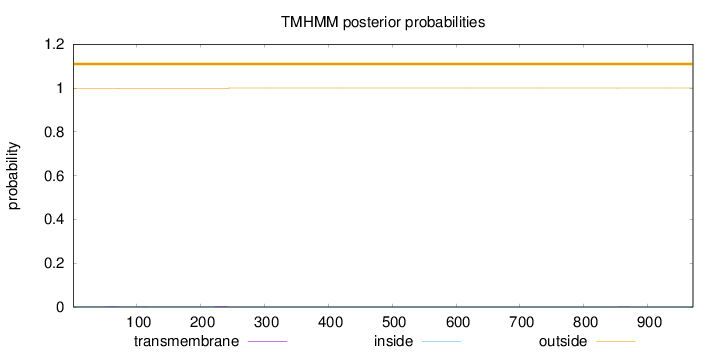
Length:
971
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0733599999999998
Exp number, first 60 AAs:
0.01326
Total prob of N-in:
0.00344
outside
1 - 971
Population Genetic Test Statistics
Pi
217.610528
Theta
189.978457
Tajima's D
-1.18148
CLR
0.583111
CSRT
0.103994800259987
Interpretation
Uncertain
