Gene
KWMTBOMO10542 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006977
Annotation
putative_Short-chain_specific_acyl-CoA_dehydrogenase?_mitochondrial_precursor_[Danaus_plexippus]
Location in the cell
Mitochondrial Reliability : 2.773
Sequence
CDS
ATGGCAACAGGTGTCGCACTAAAGTCTATAAGACTTAATAATTTGCTCTCGTCGTCTTGCGTGAAGGCGTTAGCGTCCCAATGCAGAAGCTTCACGAGTCAGCTGACGGAACAACAAAAAGAATATCAAGCATTGTCCAAGGAATTCGCCGACAAATACTTGAAGCCGAACGCAGCGAAGCATGACTTGGAAGGACGGTTCCCTTTTGATGTGATAAAAAAGCTGACAGCCACAGGCCTGATGGGCGCCTGCGTCGACGAAGAATACGGTGGCAAGGGTCTAGACTACTTGACACTGGCCGTCGCCGTGGAGGAGCTGTCCCGGGGCTGCGCCAGTACTGGCATGATCGCATCCATTCACAACTTCCTGTACGCGAACTTGGTGAACGAAAAGGGAACTCCCGAACAGAAGGAACTGTTCCTGAGGGACTACACTAAGGGCTCTCTAGGATGCTTCGCCCTCAGTGAACCTGGTGCAGGCACAGACGTGGCCAGTATACGAACGAGGGCGAAGAAGGACGGCGACCATTGGATTCTGAACGGCGAGAAGAGCTGGGTCACGACCGCCGTCGAGGGATCCGCTAATATCATATTCGCGACCTTTGACCCCGAGCTCAAGCACAAAGGAATAGCTTGTTTCCTTGTCCCTATGGACGTCGATGGCGTATTCAGAGGACGGAAAGAGGCCATAATGAGTGTCAGGGCCGTGACGTCATGCGCCCTGACCCTCGAAGACGTGAGGGTGCCGCAGAGCTACCTGGTCGGGGAGCCGGGCGAAGGGTTCAAGATCGCCATGGAGCAGCTGGACCAGGGACGGATTGGGATCGCCGCCCATGCTGTGGGAATAGCACAGGCGGCTCTGGACACCGCGATCAATTACGCCAAAGAGAGACAGGCGTTTGGGAAGCCCCTCTCCCGGTTGCCTTCCGTCAAGGATCGTTTGACGGACATGTGCATACTGGTGGAGACGGCTCGGCTGGCGACATACAGGGCGGCGACCGAGGTCAGCACCAAGAACAGCGCCATGGCGAAGTACATCGCGGGCCGGAACGCGGCCGCCGTGGCCGACCACTGCGTGCAGATCTTGGGGGGCAAGGGACTCTCCACACGATACTCGGCTGAGAGACATTACAGGGATGCCAGAGGTACTCAAATATACGGAGGAGTCACGGACATACAGAAGCGACTAGTTGGTCACTTCCTTCTGAAAGAGCACGACGCGCTATGA
Protein
MATGVALKSIRLNNLLSSSCVKALASQCRSFTSQLTEQQKEYQALSKEFADKYLKPNAAKHDLEGRFPFDVIKKLTATGLMGACVDEEYGGKGLDYLTLAVAVEELSRGCASTGMIASIHNFLYANLVNEKGTPEQKELFLRDYTKGSLGCFALSEPGAGTDVASIRTRAKKDGDHWILNGEKSWVTTAVEGSANIIFATFDPELKHKGIACFLVPMDVDGVFRGRKEAIMSVRAVTSCALTLEDVRVPQSYLVGEPGEGFKIAMEQLDQGRIGIAAHAVGIAQAALDTAINYAKERQAFGKPLSRLPSVKDRLTDMCILVETARLATYRAATEVSTKNSAMAKYIAGRNAAAVADHCVQILGGKGLSTRYSAERHYRDARGTQIYGGVTDIQKRLVGHFLLKEHDAL
Summary
Cofactor
FAD
Similarity
Belongs to the acyl-CoA dehydrogenase family.
Uniprot
H9JBT2
A0A2H1VWP4
A0A212EI46
A0A2A4JTN9
A0A194PJB9
A0A194RJN5
+ More
U5ETE1 A0A1W4WE21 A0A067QRN4 A0A2J7RDZ8 Q178X6 A0A182MIJ7 A0A1Q3FIW8 A0A182U1T9 A0A182X5Z2 A0A0P6IYW3 A0A182YQH9 D6WXB7 A0A1S4GYQ2 Q7Q8J0 A0A182RK82 A0A1J1I296 A0A182KMF8 A0A182IBM4 A0A182UZW0 A0A182WFJ6 A0A182KFN6 A0A182P4B5 A0A182QMC9 A0A068FL57 H3BF73 A0A091IGT6 A0A3L8S6H2 A0A1J1J5M8 A0A182NTC8 F1PLG8 F1PZX6 A0A3S2LN21 A0A084VFF0 A0A210PXG2 A0A3Q7U1V0 H0ZH79 U3JYI6 S4P5F9 A0A2U3Y2I2 H9JUC7 A0A091ELE1 A0A2M4BR67 A0A1Y1KIB4 A0A3M0K425 A0A2Y9I5F2 A0A1L8E2B4 A0A1L8E378 T1GCW4 A0A2Y9KUZ4 H0VTB8 A0A182Q2T6 M1EFI7 A0A2M3ZHR9 A0A218V3B7 M3XVY8 A0A182F884 U6DB06 C1C4G0 A0A250YHT4 A0A2M3ZI48 A0A2M4BPU5 A0A2H1V0Y7 A0A2M3ZI12 Q5ZL56 A0A1B0D978 A0A2E6VVC6 A0A3P4SAI7 A0A336MX34 A0A2A4K8V0 A0A3Q7RX23 A0A2M4BR37 A0A226P0H1 D2I154 Q6P324 A0A3Q7X8Z0 B0WUQ6 A0A182JR73 A0A151N4Z1 G1LPC8 A0A1B0GIU4 M3VYF9 A0A0K8TN25 A0A2U3VNP3 A0A091S4N4 Q294T1 B4GMN1 A0A1Y9H2B2 V9KL54 I3N2U4 A0A1A6HQR2 A0A093Q1H8 Q7T0Y5
U5ETE1 A0A1W4WE21 A0A067QRN4 A0A2J7RDZ8 Q178X6 A0A182MIJ7 A0A1Q3FIW8 A0A182U1T9 A0A182X5Z2 A0A0P6IYW3 A0A182YQH9 D6WXB7 A0A1S4GYQ2 Q7Q8J0 A0A182RK82 A0A1J1I296 A0A182KMF8 A0A182IBM4 A0A182UZW0 A0A182WFJ6 A0A182KFN6 A0A182P4B5 A0A182QMC9 A0A068FL57 H3BF73 A0A091IGT6 A0A3L8S6H2 A0A1J1J5M8 A0A182NTC8 F1PLG8 F1PZX6 A0A3S2LN21 A0A084VFF0 A0A210PXG2 A0A3Q7U1V0 H0ZH79 U3JYI6 S4P5F9 A0A2U3Y2I2 H9JUC7 A0A091ELE1 A0A2M4BR67 A0A1Y1KIB4 A0A3M0K425 A0A2Y9I5F2 A0A1L8E2B4 A0A1L8E378 T1GCW4 A0A2Y9KUZ4 H0VTB8 A0A182Q2T6 M1EFI7 A0A2M3ZHR9 A0A218V3B7 M3XVY8 A0A182F884 U6DB06 C1C4G0 A0A250YHT4 A0A2M3ZI48 A0A2M4BPU5 A0A2H1V0Y7 A0A2M3ZI12 Q5ZL56 A0A1B0D978 A0A2E6VVC6 A0A3P4SAI7 A0A336MX34 A0A2A4K8V0 A0A3Q7RX23 A0A2M4BR37 A0A226P0H1 D2I154 Q6P324 A0A3Q7X8Z0 B0WUQ6 A0A182JR73 A0A151N4Z1 G1LPC8 A0A1B0GIU4 M3VYF9 A0A0K8TN25 A0A2U3VNP3 A0A091S4N4 Q294T1 B4GMN1 A0A1Y9H2B2 V9KL54 I3N2U4 A0A1A6HQR2 A0A093Q1H8 Q7T0Y5
Pubmed
19121390
22118469
26354079
24845553
17510324
26999592
+ More
25244985 18362917 19820115 12364791 20966253 26385554 9215903 30282656 16341006 24438588 28812685 20360741 23622113 28004739 21993624 23236062 28087693 15592404 15642098 29337314 24621616 20010809 20431018 22293439 17975172 26369729 15632085 17994087 24402279 27762356
25244985 18362917 19820115 12364791 20966253 26385554 9215903 30282656 16341006 24438588 28812685 20360741 23622113 28004739 21993624 23236062 28087693 15592404 15642098 29337314 24621616 20010809 20431018 22293439 17975172 26369729 15632085 17994087 24402279 27762356
EMBL
BABH01015092
BABH01015093
BABH01015094
ODYU01004896
SOQ45245.1
AGBW02014713
+ More
OWR41141.1 NWSH01000689 PCG74770.1 KQ459603 KPI93133.1 KQ460124 KPJ17649.1 GANO01002825 JAB57046.1 KK853408 KDR07800.1 NEVH01005280 PNF39059.1 CH477357 EAT42766.1 AXCM01005597 GFDL01007577 JAV27468.1 GDUN01000885 JAN95034.1 KQ971361 EFA08007.1 AAAB01008944 EAA10182.4 CVRI01000038 CRK94413.1 APCN01000710 AXCN02000715 KJ622078 AID66668.1 AFYH01006469 AFYH01006470 AFYH01006471 KL218758 KFP07824.1 QUSF01000052 RLV97765.1 CVRI01000070 CRL07274.1 AAEX03014741 RSAL01000051 RVE50229.1 ATLV01012386 KE524787 KFB36694.1 NEDP02005420 OWF41152.1 ABQF01029530 ABQF01029531 AGTO01000187 GAIX01005569 JAA86991.1 BABH01038556 KK718703 KFO57881.1 GGFJ01006353 MBW55494.1 GEZM01086635 JAV59136.1 QRBI01000121 RMC05974.1 GFDF01001201 JAV12883.1 GFDF01001139 JAV12945.1 CAQQ02146330 CAQQ02146331 AAKN02007158 AXCN02000031 JP004995 AER93592.1 GGFM01007353 MBW28104.1 MUZQ01000064 OWK60221.1 AEYP01033950 HAAF01007982 CCP79806.1 BT081739 ACO51870.1 GFFW01001699 JAV43089.1 GGFM01007394 MBW28145.1 GGFJ01005949 MBW55090.1 ODYU01000178 SOQ34523.1 GGFM01007401 MBW28152.1 AADN05000505 AC160646 AJ719878 CAG31537.1 AJVK01004271 PBLQ01000055 MBL92899.1 CYRY02047052 VCX42928.1 UFQS01002855 UFQT01002855 SSX14769.1 SSX34161.1 NWSH01000035 PCG80426.1 GGFJ01006323 MBW55464.1 AWGT02000222 OXB73321.1 GL193954 EFB23137.1 AAMC01008483 AAMC01008484 BC064210 CR848265 AAH64210.1 CAJ83244.1 DS232108 EDS35042.1 AKHW03004053 KYO31685.1 ACTA01052563 AJWK01017657 AANG04002472 GDAI01002050 JAI15553.1 KK941339 KFQ50934.1 CM000070 EAL28883.2 CH479185 EDW38105.1 JW866404 AFO98921.1 AGTP01090458 LZPO01017311 OBS80798.1 KL671413 KFW82501.1 BC055986 BC084756 CM004466 AAH55986.1 AAH84756.1 OCU01793.1
OWR41141.1 NWSH01000689 PCG74770.1 KQ459603 KPI93133.1 KQ460124 KPJ17649.1 GANO01002825 JAB57046.1 KK853408 KDR07800.1 NEVH01005280 PNF39059.1 CH477357 EAT42766.1 AXCM01005597 GFDL01007577 JAV27468.1 GDUN01000885 JAN95034.1 KQ971361 EFA08007.1 AAAB01008944 EAA10182.4 CVRI01000038 CRK94413.1 APCN01000710 AXCN02000715 KJ622078 AID66668.1 AFYH01006469 AFYH01006470 AFYH01006471 KL218758 KFP07824.1 QUSF01000052 RLV97765.1 CVRI01000070 CRL07274.1 AAEX03014741 RSAL01000051 RVE50229.1 ATLV01012386 KE524787 KFB36694.1 NEDP02005420 OWF41152.1 ABQF01029530 ABQF01029531 AGTO01000187 GAIX01005569 JAA86991.1 BABH01038556 KK718703 KFO57881.1 GGFJ01006353 MBW55494.1 GEZM01086635 JAV59136.1 QRBI01000121 RMC05974.1 GFDF01001201 JAV12883.1 GFDF01001139 JAV12945.1 CAQQ02146330 CAQQ02146331 AAKN02007158 AXCN02000031 JP004995 AER93592.1 GGFM01007353 MBW28104.1 MUZQ01000064 OWK60221.1 AEYP01033950 HAAF01007982 CCP79806.1 BT081739 ACO51870.1 GFFW01001699 JAV43089.1 GGFM01007394 MBW28145.1 GGFJ01005949 MBW55090.1 ODYU01000178 SOQ34523.1 GGFM01007401 MBW28152.1 AADN05000505 AC160646 AJ719878 CAG31537.1 AJVK01004271 PBLQ01000055 MBL92899.1 CYRY02047052 VCX42928.1 UFQS01002855 UFQT01002855 SSX14769.1 SSX34161.1 NWSH01000035 PCG80426.1 GGFJ01006323 MBW55464.1 AWGT02000222 OXB73321.1 GL193954 EFB23137.1 AAMC01008483 AAMC01008484 BC064210 CR848265 AAH64210.1 CAJ83244.1 DS232108 EDS35042.1 AKHW03004053 KYO31685.1 ACTA01052563 AJWK01017657 AANG04002472 GDAI01002050 JAI15553.1 KK941339 KFQ50934.1 CM000070 EAL28883.2 CH479185 EDW38105.1 JW866404 AFO98921.1 AGTP01090458 LZPO01017311 OBS80798.1 KL671413 KFW82501.1 BC055986 BC084756 CM004466 AAH55986.1 AAH84756.1 OCU01793.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053268
UP000053240
UP000192223
+ More
UP000027135 UP000235965 UP000008820 UP000075883 UP000075902 UP000076407 UP000076408 UP000007266 UP000007062 UP000075900 UP000183832 UP000075882 UP000075840 UP000075903 UP000075920 UP000075881 UP000075885 UP000075886 UP000008672 UP000054308 UP000276834 UP000075884 UP000002254 UP000283053 UP000030765 UP000242188 UP000286640 UP000007754 UP000016665 UP000245341 UP000052976 UP000269221 UP000248481 UP000015102 UP000248482 UP000005447 UP000197619 UP000000715 UP000069272 UP000000539 UP000092462 UP000286641 UP000198419 UP000008143 UP000286642 UP000002320 UP000050525 UP000008912 UP000092461 UP000011712 UP000245340 UP000001819 UP000008744 UP000005215 UP000092124 UP000053258 UP000186698
UP000027135 UP000235965 UP000008820 UP000075883 UP000075902 UP000076407 UP000076408 UP000007266 UP000007062 UP000075900 UP000183832 UP000075882 UP000075840 UP000075903 UP000075920 UP000075881 UP000075885 UP000075886 UP000008672 UP000054308 UP000276834 UP000075884 UP000002254 UP000283053 UP000030765 UP000242188 UP000286640 UP000007754 UP000016665 UP000245341 UP000052976 UP000269221 UP000248481 UP000015102 UP000248482 UP000005447 UP000197619 UP000000715 UP000069272 UP000000539 UP000092462 UP000286641 UP000198419 UP000008143 UP000286642 UP000002320 UP000050525 UP000008912 UP000092461 UP000011712 UP000245340 UP000001819 UP000008744 UP000005215 UP000092124 UP000053258 UP000186698
Pfam
Interpro
IPR006089
Acyl-CoA_DH_CS
+ More
IPR013786 AcylCoA_DH/ox_N
IPR037069 AcylCoA_DH/ox_N_sf
IPR036250 AcylCo_DH-like_C
IPR006091 Acyl-CoA_Oxase/DH_cen-dom
IPR009100 AcylCoA_DH/oxidase_NM_dom
IPR009075 AcylCo_DH/oxidase_C
IPR026906 LRR_5
IPR003591 Leu-rich_rpt_typical-subtyp
IPR001611 Leu-rich_rpt
IPR032675 LRR_dom_sf
IPR013786 AcylCoA_DH/ox_N
IPR037069 AcylCoA_DH/ox_N_sf
IPR036250 AcylCo_DH-like_C
IPR006091 Acyl-CoA_Oxase/DH_cen-dom
IPR009100 AcylCoA_DH/oxidase_NM_dom
IPR009075 AcylCo_DH/oxidase_C
IPR026906 LRR_5
IPR003591 Leu-rich_rpt_typical-subtyp
IPR001611 Leu-rich_rpt
IPR032675 LRR_dom_sf
Gene 3D
ProteinModelPortal
H9JBT2
A0A2H1VWP4
A0A212EI46
A0A2A4JTN9
A0A194PJB9
A0A194RJN5
+ More
U5ETE1 A0A1W4WE21 A0A067QRN4 A0A2J7RDZ8 Q178X6 A0A182MIJ7 A0A1Q3FIW8 A0A182U1T9 A0A182X5Z2 A0A0P6IYW3 A0A182YQH9 D6WXB7 A0A1S4GYQ2 Q7Q8J0 A0A182RK82 A0A1J1I296 A0A182KMF8 A0A182IBM4 A0A182UZW0 A0A182WFJ6 A0A182KFN6 A0A182P4B5 A0A182QMC9 A0A068FL57 H3BF73 A0A091IGT6 A0A3L8S6H2 A0A1J1J5M8 A0A182NTC8 F1PLG8 F1PZX6 A0A3S2LN21 A0A084VFF0 A0A210PXG2 A0A3Q7U1V0 H0ZH79 U3JYI6 S4P5F9 A0A2U3Y2I2 H9JUC7 A0A091ELE1 A0A2M4BR67 A0A1Y1KIB4 A0A3M0K425 A0A2Y9I5F2 A0A1L8E2B4 A0A1L8E378 T1GCW4 A0A2Y9KUZ4 H0VTB8 A0A182Q2T6 M1EFI7 A0A2M3ZHR9 A0A218V3B7 M3XVY8 A0A182F884 U6DB06 C1C4G0 A0A250YHT4 A0A2M3ZI48 A0A2M4BPU5 A0A2H1V0Y7 A0A2M3ZI12 Q5ZL56 A0A1B0D978 A0A2E6VVC6 A0A3P4SAI7 A0A336MX34 A0A2A4K8V0 A0A3Q7RX23 A0A2M4BR37 A0A226P0H1 D2I154 Q6P324 A0A3Q7X8Z0 B0WUQ6 A0A182JR73 A0A151N4Z1 G1LPC8 A0A1B0GIU4 M3VYF9 A0A0K8TN25 A0A2U3VNP3 A0A091S4N4 Q294T1 B4GMN1 A0A1Y9H2B2 V9KL54 I3N2U4 A0A1A6HQR2 A0A093Q1H8 Q7T0Y5
U5ETE1 A0A1W4WE21 A0A067QRN4 A0A2J7RDZ8 Q178X6 A0A182MIJ7 A0A1Q3FIW8 A0A182U1T9 A0A182X5Z2 A0A0P6IYW3 A0A182YQH9 D6WXB7 A0A1S4GYQ2 Q7Q8J0 A0A182RK82 A0A1J1I296 A0A182KMF8 A0A182IBM4 A0A182UZW0 A0A182WFJ6 A0A182KFN6 A0A182P4B5 A0A182QMC9 A0A068FL57 H3BF73 A0A091IGT6 A0A3L8S6H2 A0A1J1J5M8 A0A182NTC8 F1PLG8 F1PZX6 A0A3S2LN21 A0A084VFF0 A0A210PXG2 A0A3Q7U1V0 H0ZH79 U3JYI6 S4P5F9 A0A2U3Y2I2 H9JUC7 A0A091ELE1 A0A2M4BR67 A0A1Y1KIB4 A0A3M0K425 A0A2Y9I5F2 A0A1L8E2B4 A0A1L8E378 T1GCW4 A0A2Y9KUZ4 H0VTB8 A0A182Q2T6 M1EFI7 A0A2M3ZHR9 A0A218V3B7 M3XVY8 A0A182F884 U6DB06 C1C4G0 A0A250YHT4 A0A2M3ZI48 A0A2M4BPU5 A0A2H1V0Y7 A0A2M3ZI12 Q5ZL56 A0A1B0D978 A0A2E6VVC6 A0A3P4SAI7 A0A336MX34 A0A2A4K8V0 A0A3Q7RX23 A0A2M4BR37 A0A226P0H1 D2I154 Q6P324 A0A3Q7X8Z0 B0WUQ6 A0A182JR73 A0A151N4Z1 G1LPC8 A0A1B0GIU4 M3VYF9 A0A0K8TN25 A0A2U3VNP3 A0A091S4N4 Q294T1 B4GMN1 A0A1Y9H2B2 V9KL54 I3N2U4 A0A1A6HQR2 A0A093Q1H8 Q7T0Y5
PDB
1JQI
E-value=1.54776e-85,
Score=806
Ontologies
KEGG
GO
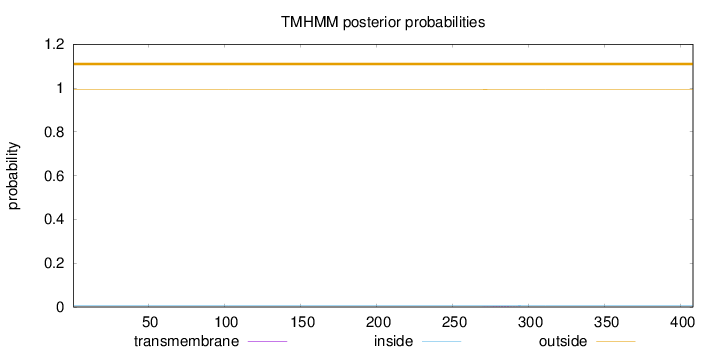
Topology
Length:
408
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03151
Exp number, first 60 AAs:
0.00058
Total prob of N-in:
0.00585
outside
1 - 408
Population Genetic Test Statistics
Pi
30.050296
Theta
167.756658
Tajima's D
-1.921934
CLR
1675.426321
CSRT
0.0173991300434978
Interpretation
Possibly Positive selection
