Gene
KWMTBOMO10541 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006978
Annotation
Leucine-rich_repeat-containing_G-protein_coupled_receptor_5_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 2.384
Sequence
CDS
ATGCTTCTAAAATATTTTTTGAGTTTTTTATTTTTATTCAAACACGTAATCGGCCAGGACGAATTGGTGACTGAATCCGTATTGAATTTGAAACAAGTGTGCGATTATTGTACGTGCAGTGAGATATCGGATATAGAGAGTTCGCATTTAGTGTTGAACATATTGTGTTCGGAGTTTGACAGGACCGCGGAACTGTCCGATCTGGACAAAATCGCTTGGCCTCCAAATCCGAACGGACTGAAGATATCCGCTTCGTTTGAAGGGATCGGACTCAGTACACTTGGCAAATTACCGCCAAATTCGCAAGTGGAAACATTAAAATTTATCAACAATGGCATCAAGACGTACTGGCCAGATCCATTTAGCGACACGCCAACCCTCAAGCGACTATCGTTATCGCAAAACGACCTAGTAGAGGTCACGCCCGACCTATTCACAAACATCGACTACCTGGAAGAACTGGATTTGTCTTACAACAAATTACAGAATTTCAATCCTTTCGATTTTAAGCATCTCAAGCATTTAAAGCGATTGAATTTGCAAAGTAATCAGATAAAGAAAATACCGGTTGAAGCGCTGCAGCCGTTGGTGGCGCTTGAAGATTTAGATTTGAGCAAAAATGGAATTTACGATTTGATACTGCCGAAAGATACAAATGCGCTTTCTGGATTGAAGAGGCTTTCTTTGAGCGGAAATAGAATTCGATCAATAGTGAAGAGTTCTTTTCCTGTCGATAATAATATAGAACTTCTGGACTTATCGAACAATATAATAGAAATAGTTGAAGAGGACTCTTTTCTATCGTTTTCCAACTTAAGAGAACTTAATCTTGGTCAGAACAACATTACCCTAGTGTTTTCATTGCCAGTAACACTGCAGATTGCCATACTAAAGATAAATACGTTGTACCATTGGCCGAAATTTCCTGTTGGTATCAAATATATTGATTTGTCGTACAATAGACTGTCGGAAGTGTATGATGAGAGCGTTGTTAATTTCGAAAGTTTAGAGGTTCTAAACATCGGCGGTAATCAAATCAAAGAAATGAATATAGAAAAGAAGTTACCGAATTTATACACGTTAGACCTATCATACAATCTAATAGCGGAAATACCAAAGACGCTGAATGCGCAGTTTTTCCCTAATTTAGACGAATTACGTTTAGATGGCAACCCGATCGAGACGGTTTATTTTAAAAATATATTGGCTTTAAAACGTTTGTATATGAATGAGTTGAATAAATTGGTTGTGGTTGATGAGAAAGCTTTTAGTAATGTGGTGGGTCGAGGGATGGACGAGGGGGGAGAGGAACTGAACTGCTTCTCATTGTACTTGTCTAATTGTCCTGCTTTGACCGAGATAAAGGAGGGCGCTTTTGACGGGACCTCTTTGTGCACGCTGGACATAAGCAAGAACAACTTGAGCCATTTATCCAAGAATCTACTAGACTGGAGCATTGCGACGGAAGGCATTAATCTACAACACAATCCTTGGCACTGCTCCTGCGACATGCAATGGGTGGTGGACGATGTCCTTCCGCAACTGTACTATACCAATAACTCCAGACTCATGGACGACTTGAGGTGCGGTTCTCCCCGAGGATACGAAGGTCTCCGACTAGTGCACTGGTACAACTGGACAGCGCAAGCGATGTGCAGCGACGTGCTAGACTTGGAACGATCCGGAGCTCACGGCAATTTCATGATGGAGCCCTCGTCGGAAAGTCCTAGAGTTAGCTCCCTGACTTTAATCTTAGCGGCATGCATTATTGTATCCCTCCTCGTTGCTATAGCTCTGACAGTATACCTCGTGAAGACCAGAAAGAAATACAGAATACGACAAGCAGCAATGAAAAGGAAAAGACAAAGCGACCTAGACTTAAGACATTCGAATGGAGTGCAAAAAGAACAGTTCTCAGCGCTGAACAAAGTTTAG
Protein
MLLKYFLSFLFLFKHVIGQDELVTESVLNLKQVCDYCTCSEISDIESSHLVLNILCSEFDRTAELSDLDKIAWPPNPNGLKISASFEGIGLSTLGKLPPNSQVETLKFINNGIKTYWPDPFSDTPTLKRLSLSQNDLVEVTPDLFTNIDYLEELDLSYNKLQNFNPFDFKHLKHLKRLNLQSNQIKKIPVEALQPLVALEDLDLSKNGIYDLILPKDTNALSGLKRLSLSGNRIRSIVKSSFPVDNNIELLDLSNNIIEIVEEDSFLSFSNLRELNLGQNNITLVFSLPVTLQIAILKINTLYHWPKFPVGIKYIDLSYNRLSEVYDESVVNFESLEVLNIGGNQIKEMNIEKKLPNLYTLDLSYNLIAEIPKTLNAQFFPNLDELRLDGNPIETVYFKNILALKRLYMNELNKLVVVDEKAFSNVVGRGMDEGGEELNCFSLYLSNCPALTEIKEGAFDGTSLCTLDISKNNLSHLSKNLLDWSIATEGINLQHNPWHCSCDMQWVVDDVLPQLYYTNNSRLMDDLRCGSPRGYEGLRLVHWYNWTAQAMCSDVLDLERSGAHGNFMMEPSSESPRVSSLTLILAACIIVSLLVAIALTVYLVKTRKKYRIRQAAMKRKRQSDLDLRHSNGVQKEQFSALNKV
Summary
Uniprot
A0A2H1VYE9
A0A2A4JSG3
A0A194PII7
A0A194RJN5
A0A212EI31
A0A3S2LAS8
+ More
A0A1L8DKP9 A0A1S4FBR2 Q178X4 A0A1B0CQW7 A0A1W7R8Q5 A0A1J1J5Q2 B0WUQ8 A0A1Q3FHZ2 A0A182G219 A0A154P130 A0A088A587 A0A1B6D6Q5 A0A2A3ET66 A0A2J7RDM7 A0A0C9QCN2 A0A0C9R774 A0A1B6GM32 A0A1B6I1H0 A0A1B6L2C9 A0A026W4T7 A0A182WGK9 A0A182MPE7 C3ZL09 W4YPB3 A0A182HRV5 K1P9F1 A0A210QFY2 A0A182U194 A0A0L8GXS8
A0A1L8DKP9 A0A1S4FBR2 Q178X4 A0A1B0CQW7 A0A1W7R8Q5 A0A1J1J5Q2 B0WUQ8 A0A1Q3FHZ2 A0A182G219 A0A154P130 A0A088A587 A0A1B6D6Q5 A0A2A3ET66 A0A2J7RDM7 A0A0C9QCN2 A0A0C9R774 A0A1B6GM32 A0A1B6I1H0 A0A1B6L2C9 A0A026W4T7 A0A182WGK9 A0A182MPE7 C3ZL09 W4YPB3 A0A182HRV5 K1P9F1 A0A210QFY2 A0A182U194 A0A0L8GXS8
EMBL
ODYU01004896
SOQ45244.1
NWSH01000689
PCG74769.1
KQ459603
KPI93132.1
+ More
KQ460124 KPJ17649.1 AGBW02014713 OWR41142.1 RSAL01000062 RVE49541.1 GFDF01007062 JAV07022.1 CH477357 EAT42768.1 AJWK01024152 AJWK01024153 AJWK01024154 AJWK01024155 GEHC01000107 JAV47538.1 CVRI01000066 CRL06217.1 DS232108 EDS35044.1 GFDL01007854 JAV27191.1 JXUM01006532 KQ560217 KXJ83773.1 KQ434783 KZC04840.1 GEDC01015934 JAS21364.1 KZ288191 PBC34492.1 NEVH01005288 PNF38930.1 GBYB01012178 JAG81945.1 GBYB01012179 JAG81946.1 GECZ01023033 GECZ01019050 GECZ01018473 GECZ01006281 JAS46736.1 JAS50719.1 JAS51296.1 JAS63488.1 GECU01026972 JAS80734.1 GEBQ01022095 JAT17882.1 KK107419 EZA51072.1 AXCM01003106 GG666640 EEN46691.1 AAGJ04005476 APCN01001759 JH818836 EKC20407.1 NEDP02003828 OWF47667.1 KQ420075 KOF81395.1
KQ460124 KPJ17649.1 AGBW02014713 OWR41142.1 RSAL01000062 RVE49541.1 GFDF01007062 JAV07022.1 CH477357 EAT42768.1 AJWK01024152 AJWK01024153 AJWK01024154 AJWK01024155 GEHC01000107 JAV47538.1 CVRI01000066 CRL06217.1 DS232108 EDS35044.1 GFDL01007854 JAV27191.1 JXUM01006532 KQ560217 KXJ83773.1 KQ434783 KZC04840.1 GEDC01015934 JAS21364.1 KZ288191 PBC34492.1 NEVH01005288 PNF38930.1 GBYB01012178 JAG81945.1 GBYB01012179 JAG81946.1 GECZ01023033 GECZ01019050 GECZ01018473 GECZ01006281 JAS46736.1 JAS50719.1 JAS51296.1 JAS63488.1 GECU01026972 JAS80734.1 GEBQ01022095 JAT17882.1 KK107419 EZA51072.1 AXCM01003106 GG666640 EEN46691.1 AAGJ04005476 APCN01001759 JH818836 EKC20407.1 NEDP02003828 OWF47667.1 KQ420075 KOF81395.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000007151
UP000283053
UP000008820
+ More
UP000092461 UP000183832 UP000002320 UP000069940 UP000249989 UP000076502 UP000005203 UP000242457 UP000235965 UP000053097 UP000075920 UP000075883 UP000001554 UP000007110 UP000075840 UP000005408 UP000242188 UP000075902 UP000053454
UP000092461 UP000183832 UP000002320 UP000069940 UP000249989 UP000076502 UP000005203 UP000242457 UP000235965 UP000053097 UP000075920 UP000075883 UP000001554 UP000007110 UP000075840 UP000005408 UP000242188 UP000075902 UP000053454
Pfam
Interpro
IPR001611
Leu-rich_rpt
+ More
IPR003591 Leu-rich_rpt_typical-subtyp
IPR032675 LRR_dom_sf
IPR026906 LRR_5
IPR013786 AcylCoA_DH/ox_N
IPR006089 Acyl-CoA_DH_CS
IPR037069 AcylCoA_DH/ox_N_sf
IPR036250 AcylCo_DH-like_C
IPR006091 Acyl-CoA_Oxase/DH_cen-dom
IPR009100 AcylCoA_DH/oxidase_NM_dom
IPR009075 AcylCo_DH/oxidase_C
IPR000483 Cys-rich_flank_reg_C
IPR037750 COPS2
IPR000717 PCI_dom
IPR036390 WH_DNA-bd_sf
IPR008615 FNIP
IPR025875 Leu-rich_rpt_4
IPR009003 Peptidase_S1_PA
IPR035897 Toll_tir_struct_dom_sf
IPR000157 TIR_dom
IPR003591 Leu-rich_rpt_typical-subtyp
IPR032675 LRR_dom_sf
IPR026906 LRR_5
IPR013786 AcylCoA_DH/ox_N
IPR006089 Acyl-CoA_DH_CS
IPR037069 AcylCoA_DH/ox_N_sf
IPR036250 AcylCo_DH-like_C
IPR006091 Acyl-CoA_Oxase/DH_cen-dom
IPR009100 AcylCoA_DH/oxidase_NM_dom
IPR009075 AcylCo_DH/oxidase_C
IPR000483 Cys-rich_flank_reg_C
IPR037750 COPS2
IPR000717 PCI_dom
IPR036390 WH_DNA-bd_sf
IPR008615 FNIP
IPR025875 Leu-rich_rpt_4
IPR009003 Peptidase_S1_PA
IPR035897 Toll_tir_struct_dom_sf
IPR000157 TIR_dom
SUPFAM
Gene 3D
ProteinModelPortal
A0A2H1VYE9
A0A2A4JSG3
A0A194PII7
A0A194RJN5
A0A212EI31
A0A3S2LAS8
+ More
A0A1L8DKP9 A0A1S4FBR2 Q178X4 A0A1B0CQW7 A0A1W7R8Q5 A0A1J1J5Q2 B0WUQ8 A0A1Q3FHZ2 A0A182G219 A0A154P130 A0A088A587 A0A1B6D6Q5 A0A2A3ET66 A0A2J7RDM7 A0A0C9QCN2 A0A0C9R774 A0A1B6GM32 A0A1B6I1H0 A0A1B6L2C9 A0A026W4T7 A0A182WGK9 A0A182MPE7 C3ZL09 W4YPB3 A0A182HRV5 K1P9F1 A0A210QFY2 A0A182U194 A0A0L8GXS8
A0A1L8DKP9 A0A1S4FBR2 Q178X4 A0A1B0CQW7 A0A1W7R8Q5 A0A1J1J5Q2 B0WUQ8 A0A1Q3FHZ2 A0A182G219 A0A154P130 A0A088A587 A0A1B6D6Q5 A0A2A3ET66 A0A2J7RDM7 A0A0C9QCN2 A0A0C9R774 A0A1B6GM32 A0A1B6I1H0 A0A1B6L2C9 A0A026W4T7 A0A182WGK9 A0A182MPE7 C3ZL09 W4YPB3 A0A182HRV5 K1P9F1 A0A210QFY2 A0A182U194 A0A0L8GXS8
PDB
4KNG
E-value=2.77661e-18,
Score=228
Ontologies
KEGG
GO
PANTHER
Topology
Subcellular location
Cytoplasm
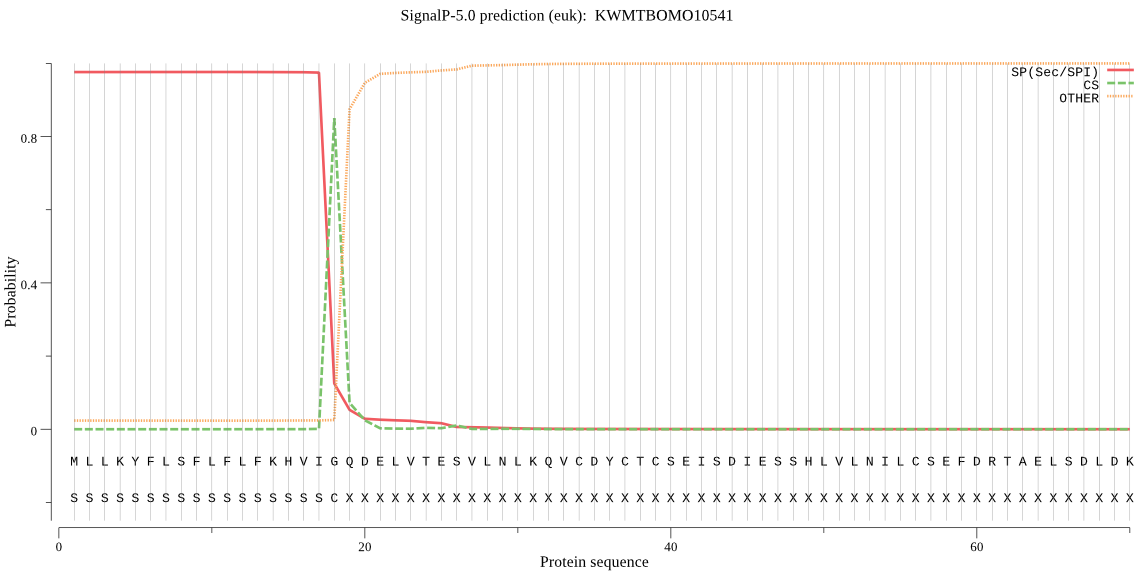
SignalP
Position: 1 - 18,
Likelihood: 0.975988
Length:
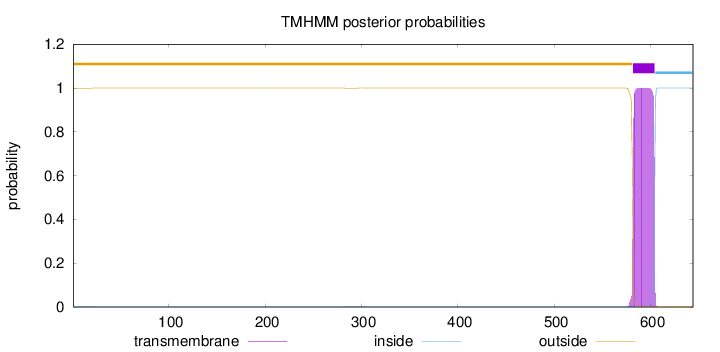
644
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.95663
Exp number, first 60 AAs:
0.00254
Total prob of N-in:
0.00046
outside
1 - 581
TMhelix
582 - 604
inside
605 - 644
Population Genetic Test Statistics
Pi
211.816312
Theta
209.422666
Tajima's D
-0.85324
CLR
46.255045
CSRT
0.167491625418729
Interpretation
Uncertain
