Gene
KWMTBOMO10540 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007136
Annotation
PREDICTED:_uncharacterized_protein_LOC106120989_[Papilio_xuthus]
Full name
Sulfide:quinone oxidoreductase, mitochondrial
Alternative Name
Sulfide quinone oxidoreductase
Sulfide dehydrogenase-like
Sulfide dehydrogenase-like
Location in the cell
Mitochondrial Reliability : 2.111
Sequence
CDS
ATGAATACAGCACGATTGTTGAGTTTTGTATCATCGTCCAACGTATGTAGAAACTTCTCAAGTTCTTCAATATGTAATGCAAATTATTCATGTAAGCTGCTAGTTGTTGGTGGAGGATCGGGTGGTTGCACAGTTGCTGCTAAATTCGCCCGTAGACTGAATAAAGACTCTGTCATTATTTTGGAACCTAGTAATGATCATTACTATCAGCCCCTTTTCACTCTGGTAGGAGCCGGAGTGAAACGTGTGAGCGACACTAGGAGATCCGCACAGAGCGTGTTACCTAAAGCAGCGAAATGGCTGAGAGATTCCGCCGAAACGATTAACGCCAAAGCGAATGTTGTGACGACGAAAGATGGACACGTCATCAATTACGAGTACATCGTTATCGCTGTTGGACTGAAGAGCGATTATAATAAAATTCCAGGCTTGACTGAGGCGCTAAACGACACTAGCAGCGGCGTGTCTACGATATATGCGCCGGAGTACTGCGAGAAGACCTGGTCTGACCTGCAGATGTTCAAGGGCGGCGAGGCCATCTTCACTTACCCGGAGACGCTGATCAAGTGTCCAGGAGCGCCGCAGAAAATTGCCTATTTAGCTGAGTCTTATTTTACCAAGACGAATGTTCGTTCGAAATCGAACATAACGTACAACACGTGCCTGCCCGTTATCTTCGGAGTGAAGAAGTACGCCGACGCGCTCCTGAAGGTGGTGGAACGGAAGAAGATCAACGTCAATTACAAGACGGTGCTCAAAGAAGTCAAACCAGACAGGAGAGAAGCGGTTTTCTACAACACTGACGACAAGACTAAGGAAACAACAATGCAATACAACATGCTCCACGTGACTCCGCCGATGAAGACCCCGGATTTCCTGCAGCGGAGTAGCGACATTGTGGACCCTAATGGCTACCTGACCGTCAACAAATATACACTGCAGCACGACAAATATCCCAACGTTTACGGCATCGGAGACTGTACCAACACTCCGAACAGCAAGACCGCCGCTGCCATTGCCAAACAATGCTACGTGCTCGAACACAATTTACTTGGTACGATGCAAAAAGATGAGCCGAAGTTAAAATACGACGGGTACGGAGCGTGCCCCCTCCTCACAGCGTACGGGAAGTGTATACTGGCCGAGTTCATTTACGACGGAATACCGCATGAAACCTTGCCCTTCGATCAGGCCCGAGAGAGCGCGTTAGCGTATTACATGAAGAAGGATCTATTCCCCTTCCTATACTGGAACTTGATGCTCAAAGGTCGTTATCACGGACCGGAATTTGTGAGAAAAATTATAAACCCATTGGCTAAGTAA
Protein
MNTARLLSFVSSSNVCRNFSSSSICNANYSCKLLVVGGGSGGCTVAAKFARRLNKDSVIILEPSNDHYYQPLFTLVGAGVKRVSDTRRSAQSVLPKAAKWLRDSAETINAKANVVTTKDGHVINYEYIVIAVGLKSDYNKIPGLTEALNDTSSGVSTIYAPEYCEKTWSDLQMFKGGEAIFTYPETLIKCPGAPQKIAYLAESYFTKTNVRSKSNITYNTCLPVIFGVKKYADALLKVVERKKINVNYKTVLKEVKPDRREAVFYNTDDKTKETTMQYNMLHVTPPMKTPDFLQRSSDIVDPNGYLTVNKYTLQHDKYPNVYGIGDCTNTPNSKTAAAIAKQCYVLEHNLLGTMQKDEPKLKYDGYGACPLLTAYGKCILAEFIYDGIPHETLPFDQARESALAYYMKKDLFPFLYWNLMLKGRYHGPEFVRKIINPLAK
Summary
Description
Catalyzes the oxidation of hydrogen sulfide with the help of a quinone, such as ubiquinone, giving rise to thiosulfate and ultimately to sulfane (molecular sulfur) atoms. Requires an additional electron acceptor; can use sulfite, sulfide or cyanide (in vitro).
Catalytic Activity
2 H(+) + hydrogen sulfide + sulfite + ubiquinone-10 = thiosulfate + ubiquinol-10
Cofactor
FAD
Similarity
Belongs to the SQRD family.
Keywords
Acetylation
Complete proteome
FAD
Flavoprotein
Mitochondrion
Oxidoreductase
Phosphoprotein
Quinone
Reference proteome
Transit peptide
3D-structure
Polymorphism
Feature
chain Sulfide:quinone oxidoreductase, mitochondrial
sequence variant In dbSNP:rs1044032.
sequence variant In dbSNP:rs1044032.
Uniprot
H9JC91
A0A2A4JSL8
A0A212EI09
A0A0N1PIL3
A0A1Y1MLF7
A0A2H1VBR8
+ More
D6WBW8 A0A182MF88 A0A2S2NC75 K7ITL0 J9K3J1 A0A1B6EDK7 A0A2Y9HII5 A0A182Q5Z9 A0A2U3X809 E9GXZ9 A0A2P2HYF4 A0A2H8TZS5 A0A182RYB8 A0A2M4AN29 C1BVL2 A0A2M4ANA4 A0A3Q2CNB5 B0WB42 A0A1Q3FHA1 A0A026W0M0 G3T2B9 B0BMT9 U5ER07 A0A1I8Q6U3 A0A182J8G8 A0A182K873 A0A3Q7WMN3 A0A3L8DN89 A0A3B3XGH5 A0A3B3U2B8 M3YIB5 A0A0K8U3B6 A0A0K8U8S2 G9KR37 A0A2U3ZP87 A0A084VZY0 A0A3Q4GBD8 A0A1L8E0E0 A0A3Q7S5U2 H2TPY3 G1M1P3 A0A182PTM6 A0A2Y9L9M2 A0A3P9PNL2 E2QTQ8 I3JNW4 A0A3Q7RHH5 A0A182HJ34 Q7QBJ7 A0A1Y9IRX1 U6CRH4 A0A286ZL94 F6Y205 A0A3Q3CHS0 A0A384BI12 B2R7T6 A0A3B4GZ10 A0A3P8NXK9 A0A3P9B7C1 A0A087XD32 A0A182XVJ5 A0A2M4D198 G1P4X2 A0A3B3C5G1 A0A3Q1JZ42 A0A1S3EP62 A0A3P9H957 H2LK25 A0A1L8E0V8 G3HMD2 A0A2K6EYL3 H2TPY4 W5JBD9 Q9R112 A0A3P9KR36 M3ZZ68 A0A1S4F0P9 A0A2M4CQ95 K7AI60 G1T782 A0A3S2MB41 G3NSH9 A0A0A1WIQ4 G3NSH3 A0A146P8J3 A0A3Q1IMN3 A0A3P8W6E5 H2Q9D6 A0A2R9CMM4 M3W7Y3 A0A1U7U9S9 A0A024R5X2 Q9Y6N5 A0A3P8U9Y8
D6WBW8 A0A182MF88 A0A2S2NC75 K7ITL0 J9K3J1 A0A1B6EDK7 A0A2Y9HII5 A0A182Q5Z9 A0A2U3X809 E9GXZ9 A0A2P2HYF4 A0A2H8TZS5 A0A182RYB8 A0A2M4AN29 C1BVL2 A0A2M4ANA4 A0A3Q2CNB5 B0WB42 A0A1Q3FHA1 A0A026W0M0 G3T2B9 B0BMT9 U5ER07 A0A1I8Q6U3 A0A182J8G8 A0A182K873 A0A3Q7WMN3 A0A3L8DN89 A0A3B3XGH5 A0A3B3U2B8 M3YIB5 A0A0K8U3B6 A0A0K8U8S2 G9KR37 A0A2U3ZP87 A0A084VZY0 A0A3Q4GBD8 A0A1L8E0E0 A0A3Q7S5U2 H2TPY3 G1M1P3 A0A182PTM6 A0A2Y9L9M2 A0A3P9PNL2 E2QTQ8 I3JNW4 A0A3Q7RHH5 A0A182HJ34 Q7QBJ7 A0A1Y9IRX1 U6CRH4 A0A286ZL94 F6Y205 A0A3Q3CHS0 A0A384BI12 B2R7T6 A0A3B4GZ10 A0A3P8NXK9 A0A3P9B7C1 A0A087XD32 A0A182XVJ5 A0A2M4D198 G1P4X2 A0A3B3C5G1 A0A3Q1JZ42 A0A1S3EP62 A0A3P9H957 H2LK25 A0A1L8E0V8 G3HMD2 A0A2K6EYL3 H2TPY4 W5JBD9 Q9R112 A0A3P9KR36 M3ZZ68 A0A1S4F0P9 A0A2M4CQ95 K7AI60 G1T782 A0A3S2MB41 G3NSH9 A0A0A1WIQ4 G3NSH3 A0A146P8J3 A0A3Q1IMN3 A0A3P8W6E5 H2Q9D6 A0A2R9CMM4 M3W7Y3 A0A1U7U9S9 A0A024R5X2 Q9Y6N5 A0A3P8U9Y8
EC Number
1.8.5.-
Pubmed
19121390
22118469
26354079
28004739
18362917
19820115
+ More
20075255 21292972 24508170 15489334 15057822 15632090 30249741 23236062 24438588 25186727 21551351 20010809 16341006 12364791 14747013 17210077 19892987 24813606 25244985 21993624 29451363 17554307 21804562 20920257 23761445 16141072 21183079 23576753 23542700 25830018 24487278 16136131 22722832 17975172 11181995 10224084 10810093 21269460 22852582 24275569 25944712
20075255 21292972 24508170 15489334 15057822 15632090 30249741 23236062 24438588 25186727 21551351 20010809 16341006 12364791 14747013 17210077 19892987 24813606 25244985 21993624 29451363 17554307 21804562 20920257 23761445 16141072 21183079 23576753 23542700 25830018 24487278 16136131 22722832 17975172 11181995 10224084 10810093 21269460 22852582 24275569 25944712
EMBL
BABH01015090
BABH01015091
BABH01015092
NWSH01000689
PCG74766.1
AGBW02014713
+ More
OWR41143.1 KQ460711 KPJ12695.1 GEZM01029788 JAV85928.1 ODYU01001709 SOQ38288.1 KQ971315 EEZ98752.1 AXCM01001682 GGMR01002195 MBY14814.1 ABLF02024989 GEDC01023111 GEDC01016060 GEDC01006153 GEDC01001269 JAS14187.1 JAS21238.1 JAS31145.1 JAS36029.1 AXCN02000238 GL732574 EFX75511.1 IACF01001087 LAB66808.1 GFXV01007971 MBW19776.1 GGFK01008874 MBW42195.1 BT078641 HACA01030284 ACO13065.1 CDW47645.1 GGFK01008891 MBW42212.1 DS231877 EDS42107.1 GFDL01008109 JAV26936.1 KK107503 EZA49605.1 AABR07053581 BC158559 CH473949 AAI58560.1 EDL80049.1 GANO01003027 JAB56844.1 QOIP01000006 RLU21349.1 AEYP01028467 AEYP01028468 AEYP01028469 AEYP01028470 AEYP01028471 AEYP01028472 AEYP01028473 GDHF01031117 JAI21197.1 GDHF01029255 JAI23059.1 JP018768 AES07366.1 ATLV01019037 KE525259 KFB43524.1 GFDF01001881 JAV12203.1 ACTA01057171 ACTA01065171 ACTA01073171 AAEX03016100 AAEX03016101 AERX01010892 APCN01002123 AAAB01008879 EAA08424.5 HAAF01000606 CCP72432.1 AEMK02000004 AK313111 BAG35933.1 AYCK01024451 GGFL01006670 MBW70848.1 AAPE02011751 AAPE02011752 AAPE02011753 GFDF01001882 JAV12202.1 JH000507 EGV93660.1 ADMH02001615 ETN61782.1 AF174535 AK008290 AK075598 AL928950 BC011153 GGFL01003322 MBW67500.1 GABC01010099 JAA01239.1 AAGW02024642 AAGW02024643 CM012442 RVE71699.1 GBXI01016039 JAC98252.1 GCES01146632 JAQ39690.1 AACZ04064526 GABF01006199 GABD01000838 GABE01005383 JAA15946.1 JAA32262.1 JAA39356.1 AJFE02111342 AJFE02111343 AJFE02111344 AANG04001761 CH471082 EAW77324.1 AF042284 AF151802 BC016836
OWR41143.1 KQ460711 KPJ12695.1 GEZM01029788 JAV85928.1 ODYU01001709 SOQ38288.1 KQ971315 EEZ98752.1 AXCM01001682 GGMR01002195 MBY14814.1 ABLF02024989 GEDC01023111 GEDC01016060 GEDC01006153 GEDC01001269 JAS14187.1 JAS21238.1 JAS31145.1 JAS36029.1 AXCN02000238 GL732574 EFX75511.1 IACF01001087 LAB66808.1 GFXV01007971 MBW19776.1 GGFK01008874 MBW42195.1 BT078641 HACA01030284 ACO13065.1 CDW47645.1 GGFK01008891 MBW42212.1 DS231877 EDS42107.1 GFDL01008109 JAV26936.1 KK107503 EZA49605.1 AABR07053581 BC158559 CH473949 AAI58560.1 EDL80049.1 GANO01003027 JAB56844.1 QOIP01000006 RLU21349.1 AEYP01028467 AEYP01028468 AEYP01028469 AEYP01028470 AEYP01028471 AEYP01028472 AEYP01028473 GDHF01031117 JAI21197.1 GDHF01029255 JAI23059.1 JP018768 AES07366.1 ATLV01019037 KE525259 KFB43524.1 GFDF01001881 JAV12203.1 ACTA01057171 ACTA01065171 ACTA01073171 AAEX03016100 AAEX03016101 AERX01010892 APCN01002123 AAAB01008879 EAA08424.5 HAAF01000606 CCP72432.1 AEMK02000004 AK313111 BAG35933.1 AYCK01024451 GGFL01006670 MBW70848.1 AAPE02011751 AAPE02011752 AAPE02011753 GFDF01001882 JAV12202.1 JH000507 EGV93660.1 ADMH02001615 ETN61782.1 AF174535 AK008290 AK075598 AL928950 BC011153 GGFL01003322 MBW67500.1 GABC01010099 JAA01239.1 AAGW02024642 AAGW02024643 CM012442 RVE71699.1 GBXI01016039 JAC98252.1 GCES01146632 JAQ39690.1 AACZ04064526 GABF01006199 GABD01000838 GABE01005383 JAA15946.1 JAA32262.1 JAA39356.1 AJFE02111342 AJFE02111343 AJFE02111344 AANG04001761 CH471082 EAW77324.1 AF042284 AF151802 BC016836
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000007266
UP000075883
+ More
UP000002358 UP000007819 UP000248481 UP000075886 UP000245341 UP000000305 UP000075900 UP000265020 UP000002320 UP000053097 UP000007646 UP000002494 UP000095300 UP000075880 UP000075881 UP000286642 UP000279307 UP000261480 UP000261500 UP000000715 UP000245340 UP000030765 UP000261580 UP000286640 UP000005226 UP000008912 UP000075885 UP000248482 UP000242638 UP000002254 UP000005207 UP000286641 UP000075840 UP000007062 UP000075903 UP000008227 UP000002281 UP000264840 UP000261680 UP000291021 UP000261460 UP000265100 UP000265160 UP000028760 UP000076408 UP000001074 UP000261560 UP000265040 UP000081671 UP000265200 UP000001038 UP000001075 UP000233160 UP000000673 UP000000589 UP000265180 UP000002852 UP000001811 UP000007635 UP000265000 UP000265120 UP000002277 UP000240080 UP000011712 UP000189704 UP000005640 UP000265080
UP000002358 UP000007819 UP000248481 UP000075886 UP000245341 UP000000305 UP000075900 UP000265020 UP000002320 UP000053097 UP000007646 UP000002494 UP000095300 UP000075880 UP000075881 UP000286642 UP000279307 UP000261480 UP000261500 UP000000715 UP000245340 UP000030765 UP000261580 UP000286640 UP000005226 UP000008912 UP000075885 UP000248482 UP000242638 UP000002254 UP000005207 UP000286641 UP000075840 UP000007062 UP000075903 UP000008227 UP000002281 UP000264840 UP000261680 UP000291021 UP000261460 UP000265100 UP000265160 UP000028760 UP000076408 UP000001074 UP000261560 UP000265040 UP000081671 UP000265200 UP000001038 UP000001075 UP000233160 UP000000673 UP000000589 UP000265180 UP000002852 UP000001811 UP000007635 UP000265000 UP000265120 UP000002277 UP000240080 UP000011712 UP000189704 UP000005640 UP000265080
Interpro
SUPFAM
SSF51905
SSF51905
Gene 3D
ProteinModelPortal
H9JC91
A0A2A4JSL8
A0A212EI09
A0A0N1PIL3
A0A1Y1MLF7
A0A2H1VBR8
+ More
D6WBW8 A0A182MF88 A0A2S2NC75 K7ITL0 J9K3J1 A0A1B6EDK7 A0A2Y9HII5 A0A182Q5Z9 A0A2U3X809 E9GXZ9 A0A2P2HYF4 A0A2H8TZS5 A0A182RYB8 A0A2M4AN29 C1BVL2 A0A2M4ANA4 A0A3Q2CNB5 B0WB42 A0A1Q3FHA1 A0A026W0M0 G3T2B9 B0BMT9 U5ER07 A0A1I8Q6U3 A0A182J8G8 A0A182K873 A0A3Q7WMN3 A0A3L8DN89 A0A3B3XGH5 A0A3B3U2B8 M3YIB5 A0A0K8U3B6 A0A0K8U8S2 G9KR37 A0A2U3ZP87 A0A084VZY0 A0A3Q4GBD8 A0A1L8E0E0 A0A3Q7S5U2 H2TPY3 G1M1P3 A0A182PTM6 A0A2Y9L9M2 A0A3P9PNL2 E2QTQ8 I3JNW4 A0A3Q7RHH5 A0A182HJ34 Q7QBJ7 A0A1Y9IRX1 U6CRH4 A0A286ZL94 F6Y205 A0A3Q3CHS0 A0A384BI12 B2R7T6 A0A3B4GZ10 A0A3P8NXK9 A0A3P9B7C1 A0A087XD32 A0A182XVJ5 A0A2M4D198 G1P4X2 A0A3B3C5G1 A0A3Q1JZ42 A0A1S3EP62 A0A3P9H957 H2LK25 A0A1L8E0V8 G3HMD2 A0A2K6EYL3 H2TPY4 W5JBD9 Q9R112 A0A3P9KR36 M3ZZ68 A0A1S4F0P9 A0A2M4CQ95 K7AI60 G1T782 A0A3S2MB41 G3NSH9 A0A0A1WIQ4 G3NSH3 A0A146P8J3 A0A3Q1IMN3 A0A3P8W6E5 H2Q9D6 A0A2R9CMM4 M3W7Y3 A0A1U7U9S9 A0A024R5X2 Q9Y6N5 A0A3P8U9Y8
D6WBW8 A0A182MF88 A0A2S2NC75 K7ITL0 J9K3J1 A0A1B6EDK7 A0A2Y9HII5 A0A182Q5Z9 A0A2U3X809 E9GXZ9 A0A2P2HYF4 A0A2H8TZS5 A0A182RYB8 A0A2M4AN29 C1BVL2 A0A2M4ANA4 A0A3Q2CNB5 B0WB42 A0A1Q3FHA1 A0A026W0M0 G3T2B9 B0BMT9 U5ER07 A0A1I8Q6U3 A0A182J8G8 A0A182K873 A0A3Q7WMN3 A0A3L8DN89 A0A3B3XGH5 A0A3B3U2B8 M3YIB5 A0A0K8U3B6 A0A0K8U8S2 G9KR37 A0A2U3ZP87 A0A084VZY0 A0A3Q4GBD8 A0A1L8E0E0 A0A3Q7S5U2 H2TPY3 G1M1P3 A0A182PTM6 A0A2Y9L9M2 A0A3P9PNL2 E2QTQ8 I3JNW4 A0A3Q7RHH5 A0A182HJ34 Q7QBJ7 A0A1Y9IRX1 U6CRH4 A0A286ZL94 F6Y205 A0A3Q3CHS0 A0A384BI12 B2R7T6 A0A3B4GZ10 A0A3P8NXK9 A0A3P9B7C1 A0A087XD32 A0A182XVJ5 A0A2M4D198 G1P4X2 A0A3B3C5G1 A0A3Q1JZ42 A0A1S3EP62 A0A3P9H957 H2LK25 A0A1L8E0V8 G3HMD2 A0A2K6EYL3 H2TPY4 W5JBD9 Q9R112 A0A3P9KR36 M3ZZ68 A0A1S4F0P9 A0A2M4CQ95 K7AI60 G1T782 A0A3S2MB41 G3NSH9 A0A0A1WIQ4 G3NSH3 A0A146P8J3 A0A3Q1IMN3 A0A3P8W6E5 H2Q9D6 A0A2R9CMM4 M3W7Y3 A0A1U7U9S9 A0A024R5X2 Q9Y6N5 A0A3P8U9Y8
PDB
6MP5
E-value=2.55962e-113,
Score=1046
Ontologies
PATHWAY
GO
PANTHER
Topology
Subcellular location
Mitochondrion
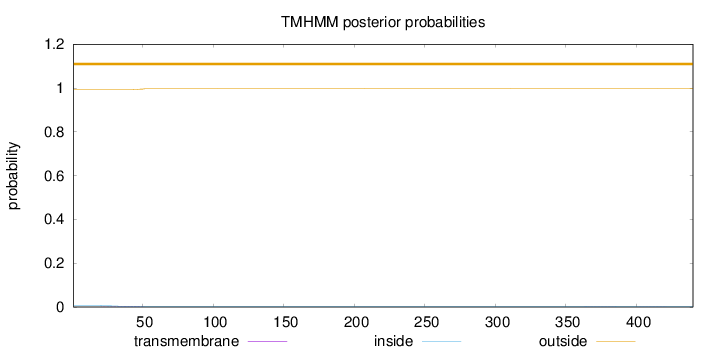
Length:
440
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.07694
Exp number, first 60 AAs:
0.06453
Total prob of N-in:
0.00617
outside
1 - 440
Population Genetic Test Statistics
Pi
24.392646
Theta
22.825293
Tajima's D
0.677308
CLR
0.000861
CSRT
0.577921103944803
Interpretation
Uncertain
