Gene
KWMTBOMO10531
Pre Gene Modal
BGIBMGA007133
Annotation
PREDICTED:_TATA-binding_protein-associated_factor_172_[Bombyx_mori]
Full name
TATA-binding protein-associated factor 172
+ More
Poly [ADP-ribose] polymerase
Poly [ADP-ribose] polymerase
Alternative Name
ATP-dependent helicase BTAF1
B-TFIID transcription factor-associated 170 kDa subunit
TAF(II)170
TBP-associated factor 172
B-TFIID transcription factor-associated 170 kDa subunit
TAF(II)170
TBP-associated factor 172
Location in the cell
Mitochondrial Reliability : 1.14
Sequence
CDS
ATGTCACTGGTGCCGGCGCTGGGCGAGGCGGGCGGGGCGCGCGTGCGGTGCGGCGCGGCGGAGGCGCTGGCGCGGCTGGTGGACGCCCTGCAGCTGCAGCTCGTGCCCTACATCGTGCTGCTCGTCATCCCGCTGCTCGGTCGTATGAGCGACCACAGCGAGGCCGTCCGCATGATGTCGACCCGTTGCTTCGCGACCCTGATCCAGCTGATGCCGCTAGAGGGCGCCGTGCCCGAGCCGCCCGACCTGGCCCCCGCGCTGCGCGACCGCATACGTCACGACAAGAACTTTCTGGAACAACTCTTCAACCCCAAATCGATCAAGGACTATAAAATCCCCGTGCCGCTCTCCGCCGAGTTACGCTCCTATCAACAGGCGGGCGTGAACTGGCTGCGGTTCCTGTACGAGTACAAGTTGCACGGCGTGCTGTGCGACGACATGGGGCTGGGCAAGACGCTGCAGTCCATAGCCGTGGTGGCGGGCTCGCACTGGGAGGGCGCCGCCGCGCGGGCGCACGCGCTGCCCTCGCTCGTCGTCTGCCCCCCCACACTCACAGGTCACTGGGTGTTCGAAGTAAACAAGTTCATACCTTCGAAGATCCTCAGGCCGCTGCAGTACGTGGGGCCGCCGGCTGAACGGGAGAAGCTTCGGCATCAAGTGAAGTACCACAACTTCATCGTCGCCTCGTACGACATCGTCAGGAAGGACATAGACTTCTTCAGCAGCATCAGGTGGAACTACTGCGTTCTGGACGAGGGACACGTTATCAAAAATGACAAGACGAAGGCTTTCAAAGCCATTAAAATGCTGATAGCCAACCATAGGTTGATACTATCCGGAACACCTATTCAGAACAACGTGCTGGAGCTGTGGTCGCTGTTCGACTTCTTGATGCCGGGCCTGCTGGGCACCTCGCGCGAGTTCACGGCGCGCTACTCGCGGCCCATCCTCGCCGCCCGCGACCCCCGCGCCTCCCCGCTGCAGCTGCAGGCCGGCGCGCTCGCCTGCGAGGCGCTGCACAGGCAGGTGCTACCGTTCCTGTTGAGGCGTGTTAAAGAAGATGTACTGAAGGAGCTTCCACCGAAGATAACGCAGGACTATTACTGCGAGCTCAGCCCCCTACAGCGATGCCTGTACGAGGACTTCTCGAGAGACCACGTGCCGCAGGACATCACTAGCACTAGTCACACACACGTATTTCAGGCTCTACATTACCTCCAAAACGTCTGCAACCACCCGAAGCTGGTGTTGACTCCGGACCATCCCGAGGCACAAACCATCGCCAAACAATTGAGAGCGCAAAATTCTAATCTCGACGACATTGAACATTCCGCTAAACTGCCTGCTTTAAAACAACTGCTCCTCGACTGCGGGATCGGCGCGACGGGGGCGGAGGCCGAGTCCTCGCCGGTCGTGTCGCAACACCGAGCGCTCGTATTCTGTCAACTGAAGAAAATGTTGGACATCGTCGAGAGAGATCTGCTGCTACAGCATCTGCCCTCCGTGACGTACCTGCGGCTCGACGGCTCGGTCCCGCCCCAGCACAGGCACGCCATCGTCACGCGCTTCAACACCGACGTCTCCATTGACTTGTTACTCCTCACGACCGCGGTGGGAGGCCTGGGGCTGAATTTAACCGGCGCGGACACTGTGATTTTCGTGGAACACGACTGGAACCCCATGAAGGACCTGCAGGCCATGGACCGAGCTCATCGGATCGGACAAAAGAAAGTTGTTAATGTCTACAGACTCATCACCAGGAATACACTCGAAGAGAAGATAATGGGGCTACAAAAATTTAAATTGATGACAGCGAACACGGTTATAAGCAGCGAGAACGCGGCCATAGAGACGATGGGCACGGACCAGCTGCTTGACCTGTTCACTCTCAAGCCCGGCCGCTCGCACGACCAGCCCGCCGCCTCCAGGTCCGTGCTGGAGGCGCTGCCCGACCTCTGGGACGACAAACAGTACGAAGAGGAATATGACATGACTAACTTCATCAAGGGATTGAACAAAATGAACGCGCACTAA
Protein
MSLVPALGEAGGARVRCGAAEALARLVDALQLQLVPYIVLLVIPLLGRMSDHSEAVRMMSTRCFATLIQLMPLEGAVPEPPDLAPALRDRIRHDKNFLEQLFNPKSIKDYKIPVPLSAELRSYQQAGVNWLRFLYEYKLHGVLCDDMGLGKTLQSIAVVAGSHWEGAAARAHALPSLVVCPPTLTGHWVFEVNKFIPSKILRPLQYVGPPAEREKLRHQVKYHNFIVASYDIVRKDIDFFSSIRWNYCVLDEGHVIKNDKTKAFKAIKMLIANHRLILSGTPIQNNVLELWSLFDFLMPGLLGTSREFTARYSRPILAARDPRASPLQLQAGALACEALHRQVLPFLLRRVKEDVLKELPPKITQDYYCELSPLQRCLYEDFSRDHVPQDITSTSHTHVFQALHYLQNVCNHPKLVLTPDHPEAQTIAKQLRAQNSNLDDIEHSAKLPALKQLLLDCGIGATGAEAESSPVVSQHRALVFCQLKKMLDIVERDLLLQHLPSVTYLRLDGSVPPQHRHAIVTRFNTDVSIDLLLLTTAVGGLGLNLTGADTVIFVEHDWNPMKDLQAMDRAHRIGQKKVVNVYRLITRNTLEEKIMGLQKFKLMTANTVISSENAAIETMGTDQLLDLFTLKPGRSHDQPAASRSVLEALPDLWDDKQYEEEYDMTNFIKGLNKMNAH
Summary
Subunit
Associates with TBP to form B-TFIID. Binds DRAP1.
Similarity
Belongs to the SNF2/RAD54 helicase family.
Keywords
Alternative splicing
ATP-binding
Complete proteome
Direct protein sequencing
DNA-binding
Helicase
Hydrolase
Nucleotide-binding
Nucleus
Phosphoprotein
Reference proteome
Repeat
Feature
chain TATA-binding protein-associated factor 172
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9JC88
A0A2A4JRU2
A0A194PII4
A0A212ERX3
A0A0N0PCB8
A0A067QSZ1
+ More
A0A1B6C462 A0A1B6DFE2 A0A2J7RIR5 A0A1Q3FBY8 A0A1Q3FES8 B0W966 A0A182GQN5 A0A182G5L0 A0A336KBP6 Q16JW5 D6WW10 A0A023F3Q0 A0A0A9X8J8 A0A224X9F8 E0VSS7 A0A1W4XGH1 A0A1J1HY29 A0A182JX29 A0A1B6CL15 A0A182IRV5 A0A182RP11 W4VRR5 A0A182XVA4 A0A2R7WMI8 A0A182HGR5 Q7PUQ9 A0A084VRP0 A0A1D2NHA0 A0A182P5N4 A0A1Y1K030 A0A0A1WE77 U4U265 N6T163 A0A182UMS2 A0A1S3HG25 A0A0K8V4N8 A0A034VBP8 A0A2K5NYE6 A0A1S3HME5 W8BMY6 A0A2R9CL81 H2Q297 A0A2J8Y586 G3S6X7 G3QWB8 A0A2R9CL47 K7CDG3 A0A0A0MTH9 Q2M1V9 O14981 A0A2K6CJ01 K7EVC3 A0A1L8DH30 A0A0G2K1N4 A0A2K6CJ03 G7PDJ8 A0A2K5VRB0 A0A096P5I6 M3Z0K8 A0A2K5IAT2 H9G1J4 A0A1L8DFY6 A0A0P6JHM9 A0A2K6CJ37 A0A2K6K815 A0A2K5VR83 A0A2K6ACZ1 A0A0D9R2N4 A0A2K5NZ06 A0A2K5NYM6 A0A2K5IAD5 A0A1D5Q9F4 A0ZVB6 E9QAE3 F1LW16 E2QWL4 A0A2K6T5A3 A0A2K6K7Z2 G3TLC7 U6DCU6 A0A2K5EAH3 A0A2K6NLM4 A0A2K6NLL1 A0A2K5IAN3 F7H9R4 A0A2K6T5E5 A0A2R8MUK8 A0A087SYZ8 A0A1S3GNE9 A0A091DCR0 A0A2Y9RVR9 A0A2K5QTX5 A0A2K5EAI7 A0A2K6NLI4
A0A1B6C462 A0A1B6DFE2 A0A2J7RIR5 A0A1Q3FBY8 A0A1Q3FES8 B0W966 A0A182GQN5 A0A182G5L0 A0A336KBP6 Q16JW5 D6WW10 A0A023F3Q0 A0A0A9X8J8 A0A224X9F8 E0VSS7 A0A1W4XGH1 A0A1J1HY29 A0A182JX29 A0A1B6CL15 A0A182IRV5 A0A182RP11 W4VRR5 A0A182XVA4 A0A2R7WMI8 A0A182HGR5 Q7PUQ9 A0A084VRP0 A0A1D2NHA0 A0A182P5N4 A0A1Y1K030 A0A0A1WE77 U4U265 N6T163 A0A182UMS2 A0A1S3HG25 A0A0K8V4N8 A0A034VBP8 A0A2K5NYE6 A0A1S3HME5 W8BMY6 A0A2R9CL81 H2Q297 A0A2J8Y586 G3S6X7 G3QWB8 A0A2R9CL47 K7CDG3 A0A0A0MTH9 Q2M1V9 O14981 A0A2K6CJ01 K7EVC3 A0A1L8DH30 A0A0G2K1N4 A0A2K6CJ03 G7PDJ8 A0A2K5VRB0 A0A096P5I6 M3Z0K8 A0A2K5IAT2 H9G1J4 A0A1L8DFY6 A0A0P6JHM9 A0A2K6CJ37 A0A2K6K815 A0A2K5VR83 A0A2K6ACZ1 A0A0D9R2N4 A0A2K5NZ06 A0A2K5NYM6 A0A2K5IAD5 A0A1D5Q9F4 A0ZVB6 E9QAE3 F1LW16 E2QWL4 A0A2K6T5A3 A0A2K6K7Z2 G3TLC7 U6DCU6 A0A2K5EAH3 A0A2K6NLM4 A0A2K6NLL1 A0A2K5IAN3 F7H9R4 A0A2K6T5E5 A0A2R8MUK8 A0A087SYZ8 A0A1S3GNE9 A0A091DCR0 A0A2Y9RVR9 A0A2K5QTX5 A0A2K5EAI7 A0A2K6NLI4
EC Number
3.6.4.-
2.4.2.-
2.4.2.-
Pubmed
19121390
26354079
22118469
24845553
26483478
17510324
+ More
18362917 19820115 25474469 25401762 20566863 25244985 12364791 14747013 17210077 24438588 27289101 28004739 25830018 23537049 25348373 24495485 22722832 16136131 22398555 15164054 21269460 23186163 11181995 15489334 9342322 9488487 14702039 10642510 15509807 15057822 16641100 22002653 17431167 25319552 19468303 21183079 16341006 25362486 25243066
18362917 19820115 25474469 25401762 20566863 25244985 12364791 14747013 17210077 24438588 27289101 28004739 25830018 23537049 25348373 24495485 22722832 16136131 22398555 15164054 21269460 23186163 11181995 15489334 9342322 9488487 14702039 10642510 15509807 15057822 16641100 22002653 17431167 25319552 19468303 21183079 16341006 25362486 25243066
EMBL
BABH01015068
BABH01015069
BABH01015070
BABH01015071
BABH01015072
BABH01015073
+ More
BABH01015074 NWSH01000689 PCG74757.1 KQ459603 KPI93127.1 AGBW02012899 OWR44242.1 KQ460711 KPJ12691.1 KK852980 KDR13008.1 GEDC01029006 JAS08292.1 GEDC01012899 JAS24399.1 NEVH01003493 PNF40724.1 GFDL01010023 JAV25022.1 GFDL01008997 JAV26048.1 DS231862 EDS39763.1 JXUM01080903 KQ563215 KXJ74274.1 JXUM01043776 JXUM01043777 JXUM01043778 UFQS01000046 UFQT01000046 SSW98500.1 SSX18886.1 CH477989 EAT34577.1 KQ971361 EFA08205.1 GBBI01003193 JAC15519.1 GBHO01026557 GBRD01014888 JAG17047.1 JAG50938.1 GFTR01008892 JAW07534.1 DS235756 EEB16433.1 CVRI01000036 CRK93000.1 GEDC01023186 GEDC01014621 JAS14112.1 JAS22677.1 GANO01000305 JAB59566.1 KK855100 PTY20838.1 APCN01005942 AAAB01008987 EAA01172.6 ATLV01015761 KE525036 KFB40634.1 LJIJ01000039 ODN04641.1 GEZM01096473 JAV54874.1 GBXI01017008 GBXI01015292 JAC97283.1 JAC98999.1 KB631617 ERL84706.1 APGK01057067 KB741277 ENN71273.1 GDHF01018412 JAI33902.1 GAKP01018256 JAC40696.1 GAMC01011820 GAMC01011819 JAB94736.1 AJFE02089207 AJFE02089208 AJFE02089209 AJFE02089210 AJFE02089211 AACZ04051054 ABGA01142724 ABGA01142725 ABGA01142726 NDHI03003280 PNJ89429.1 PNJ89430.1 CABD030075315 CABD030075316 CABD030075317 CABD030075318 CABD030075319 CABD030075320 CABD030075321 CABD030075322 CABD030075323 GABC01000314 GABF01005178 GABD01003440 GABE01005049 NBAG03000216 JAA11024.1 JAA16967.1 JAA29660.1 JAA39690.1 PNI82330.1 PNI82331.1 AL359198 AL365398 BC112201 CH471066 AAI12202.1 EAW50102.1 AJ001017 AF038362 AK303554 AF166118 GFDF01008322 JAV05762.1 AC094246 CM001284 EHH64880.1 AQIA01071978 AQIA01071979 AQIA01071980 AHZZ02035770 AHZZ02035771 AEYP01026426 AEYP01026427 AEYP01026428 AEYP01026429 AEYP01026430 AEYP01026431 AEYP01026432 AEYP01026433 AEYP01026434 AEYP01026435 JSUE03043975 JSUE03043976 JU337000 JU477728 AFE80753.1 AFH34532.1 GFDF01008716 JAV05368.1 GEBF01003065 JAO00568.1 AQIB01042875 AB287295 BAF41982.1 AC118931 AAEX03015408 HAAF01008597 CCP80421.1 GAMR01006006 GAMP01000263 JAB27926.1 JAB52492.1 KK112605 KFM58087.1 KN122536 KFO29934.1
BABH01015074 NWSH01000689 PCG74757.1 KQ459603 KPI93127.1 AGBW02012899 OWR44242.1 KQ460711 KPJ12691.1 KK852980 KDR13008.1 GEDC01029006 JAS08292.1 GEDC01012899 JAS24399.1 NEVH01003493 PNF40724.1 GFDL01010023 JAV25022.1 GFDL01008997 JAV26048.1 DS231862 EDS39763.1 JXUM01080903 KQ563215 KXJ74274.1 JXUM01043776 JXUM01043777 JXUM01043778 UFQS01000046 UFQT01000046 SSW98500.1 SSX18886.1 CH477989 EAT34577.1 KQ971361 EFA08205.1 GBBI01003193 JAC15519.1 GBHO01026557 GBRD01014888 JAG17047.1 JAG50938.1 GFTR01008892 JAW07534.1 DS235756 EEB16433.1 CVRI01000036 CRK93000.1 GEDC01023186 GEDC01014621 JAS14112.1 JAS22677.1 GANO01000305 JAB59566.1 KK855100 PTY20838.1 APCN01005942 AAAB01008987 EAA01172.6 ATLV01015761 KE525036 KFB40634.1 LJIJ01000039 ODN04641.1 GEZM01096473 JAV54874.1 GBXI01017008 GBXI01015292 JAC97283.1 JAC98999.1 KB631617 ERL84706.1 APGK01057067 KB741277 ENN71273.1 GDHF01018412 JAI33902.1 GAKP01018256 JAC40696.1 GAMC01011820 GAMC01011819 JAB94736.1 AJFE02089207 AJFE02089208 AJFE02089209 AJFE02089210 AJFE02089211 AACZ04051054 ABGA01142724 ABGA01142725 ABGA01142726 NDHI03003280 PNJ89429.1 PNJ89430.1 CABD030075315 CABD030075316 CABD030075317 CABD030075318 CABD030075319 CABD030075320 CABD030075321 CABD030075322 CABD030075323 GABC01000314 GABF01005178 GABD01003440 GABE01005049 NBAG03000216 JAA11024.1 JAA16967.1 JAA29660.1 JAA39690.1 PNI82330.1 PNI82331.1 AL359198 AL365398 BC112201 CH471066 AAI12202.1 EAW50102.1 AJ001017 AF038362 AK303554 AF166118 GFDF01008322 JAV05762.1 AC094246 CM001284 EHH64880.1 AQIA01071978 AQIA01071979 AQIA01071980 AHZZ02035770 AHZZ02035771 AEYP01026426 AEYP01026427 AEYP01026428 AEYP01026429 AEYP01026430 AEYP01026431 AEYP01026432 AEYP01026433 AEYP01026434 AEYP01026435 JSUE03043975 JSUE03043976 JU337000 JU477728 AFE80753.1 AFH34532.1 GFDF01008716 JAV05368.1 GEBF01003065 JAO00568.1 AQIB01042875 AB287295 BAF41982.1 AC118931 AAEX03015408 HAAF01008597 CCP80421.1 GAMR01006006 GAMP01000263 JAB27926.1 JAB52492.1 KK112605 KFM58087.1 KN122536 KFO29934.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000027135
+ More
UP000235965 UP000002320 UP000069940 UP000249989 UP000008820 UP000007266 UP000009046 UP000192223 UP000183832 UP000075881 UP000075880 UP000075900 UP000076408 UP000075840 UP000007062 UP000030765 UP000094527 UP000075885 UP000030742 UP000019118 UP000075903 UP000085678 UP000233060 UP000240080 UP000002277 UP000001595 UP000001519 UP000005640 UP000233120 UP000002494 UP000009130 UP000233100 UP000028761 UP000000715 UP000233080 UP000006718 UP000233180 UP000233140 UP000029965 UP000000589 UP000002254 UP000233220 UP000007646 UP000233020 UP000233200 UP000008225 UP000054359 UP000081671 UP000028990 UP000248480 UP000233040
UP000235965 UP000002320 UP000069940 UP000249989 UP000008820 UP000007266 UP000009046 UP000192223 UP000183832 UP000075881 UP000075880 UP000075900 UP000076408 UP000075840 UP000007062 UP000030765 UP000094527 UP000075885 UP000030742 UP000019118 UP000075903 UP000085678 UP000233060 UP000240080 UP000002277 UP000001595 UP000001519 UP000005640 UP000233120 UP000002494 UP000009130 UP000233100 UP000028761 UP000000715 UP000233080 UP000006718 UP000233180 UP000233140 UP000029965 UP000000589 UP000002254 UP000233220 UP000007646 UP000233020 UP000233200 UP000008225 UP000054359 UP000081671 UP000028990 UP000248480 UP000233040
Pfam
Interpro
IPR014001
Helicase_ATP-bd
+ More
IPR001650 Helicase_C
IPR016024 ARM-type_fold
IPR022707 DUF3535
IPR000330 SNF2_N
IPR027417 P-loop_NTPase
IPR038718 SNF2-like_sf
IPR011989 ARM-like
IPR001660 SAM
IPR012317 Poly(ADP-ribose)pol_cat_dom
IPR013761 SAM/pointed_sf
IPR002110 Ankyrin_rpt
IPR020683 Ankyrin_rpt-contain_dom
IPR036770 Ankyrin_rpt-contain_sf
IPR038742 FRS2_PTB
IPR002404 IRS_PTB
IPR001650 Helicase_C
IPR016024 ARM-type_fold
IPR022707 DUF3535
IPR000330 SNF2_N
IPR027417 P-loop_NTPase
IPR038718 SNF2-like_sf
IPR011989 ARM-like
IPR001660 SAM
IPR012317 Poly(ADP-ribose)pol_cat_dom
IPR013761 SAM/pointed_sf
IPR002110 Ankyrin_rpt
IPR020683 Ankyrin_rpt-contain_dom
IPR036770 Ankyrin_rpt-contain_sf
IPR038742 FRS2_PTB
IPR002404 IRS_PTB
Gene 3D
ProteinModelPortal
H9JC88
A0A2A4JRU2
A0A194PII4
A0A212ERX3
A0A0N0PCB8
A0A067QSZ1
+ More
A0A1B6C462 A0A1B6DFE2 A0A2J7RIR5 A0A1Q3FBY8 A0A1Q3FES8 B0W966 A0A182GQN5 A0A182G5L0 A0A336KBP6 Q16JW5 D6WW10 A0A023F3Q0 A0A0A9X8J8 A0A224X9F8 E0VSS7 A0A1W4XGH1 A0A1J1HY29 A0A182JX29 A0A1B6CL15 A0A182IRV5 A0A182RP11 W4VRR5 A0A182XVA4 A0A2R7WMI8 A0A182HGR5 Q7PUQ9 A0A084VRP0 A0A1D2NHA0 A0A182P5N4 A0A1Y1K030 A0A0A1WE77 U4U265 N6T163 A0A182UMS2 A0A1S3HG25 A0A0K8V4N8 A0A034VBP8 A0A2K5NYE6 A0A1S3HME5 W8BMY6 A0A2R9CL81 H2Q297 A0A2J8Y586 G3S6X7 G3QWB8 A0A2R9CL47 K7CDG3 A0A0A0MTH9 Q2M1V9 O14981 A0A2K6CJ01 K7EVC3 A0A1L8DH30 A0A0G2K1N4 A0A2K6CJ03 G7PDJ8 A0A2K5VRB0 A0A096P5I6 M3Z0K8 A0A2K5IAT2 H9G1J4 A0A1L8DFY6 A0A0P6JHM9 A0A2K6CJ37 A0A2K6K815 A0A2K5VR83 A0A2K6ACZ1 A0A0D9R2N4 A0A2K5NZ06 A0A2K5NYM6 A0A2K5IAD5 A0A1D5Q9F4 A0ZVB6 E9QAE3 F1LW16 E2QWL4 A0A2K6T5A3 A0A2K6K7Z2 G3TLC7 U6DCU6 A0A2K5EAH3 A0A2K6NLM4 A0A2K6NLL1 A0A2K5IAN3 F7H9R4 A0A2K6T5E5 A0A2R8MUK8 A0A087SYZ8 A0A1S3GNE9 A0A091DCR0 A0A2Y9RVR9 A0A2K5QTX5 A0A2K5EAI7 A0A2K6NLI4
A0A1B6C462 A0A1B6DFE2 A0A2J7RIR5 A0A1Q3FBY8 A0A1Q3FES8 B0W966 A0A182GQN5 A0A182G5L0 A0A336KBP6 Q16JW5 D6WW10 A0A023F3Q0 A0A0A9X8J8 A0A224X9F8 E0VSS7 A0A1W4XGH1 A0A1J1HY29 A0A182JX29 A0A1B6CL15 A0A182IRV5 A0A182RP11 W4VRR5 A0A182XVA4 A0A2R7WMI8 A0A182HGR5 Q7PUQ9 A0A084VRP0 A0A1D2NHA0 A0A182P5N4 A0A1Y1K030 A0A0A1WE77 U4U265 N6T163 A0A182UMS2 A0A1S3HG25 A0A0K8V4N8 A0A034VBP8 A0A2K5NYE6 A0A1S3HME5 W8BMY6 A0A2R9CL81 H2Q297 A0A2J8Y586 G3S6X7 G3QWB8 A0A2R9CL47 K7CDG3 A0A0A0MTH9 Q2M1V9 O14981 A0A2K6CJ01 K7EVC3 A0A1L8DH30 A0A0G2K1N4 A0A2K6CJ03 G7PDJ8 A0A2K5VRB0 A0A096P5I6 M3Z0K8 A0A2K5IAT2 H9G1J4 A0A1L8DFY6 A0A0P6JHM9 A0A2K6CJ37 A0A2K6K815 A0A2K5VR83 A0A2K6ACZ1 A0A0D9R2N4 A0A2K5NZ06 A0A2K5NYM6 A0A2K5IAD5 A0A1D5Q9F4 A0ZVB6 E9QAE3 F1LW16 E2QWL4 A0A2K6T5A3 A0A2K6K7Z2 G3TLC7 U6DCU6 A0A2K5EAH3 A0A2K6NLM4 A0A2K6NLL1 A0A2K5IAN3 F7H9R4 A0A2K6T5E5 A0A2R8MUK8 A0A087SYZ8 A0A1S3GNE9 A0A091DCR0 A0A2Y9RVR9 A0A2K5QTX5 A0A2K5EAI7 A0A2K6NLI4
PDB
6G7E
E-value=0,
Score=1667
Ontologies
GO
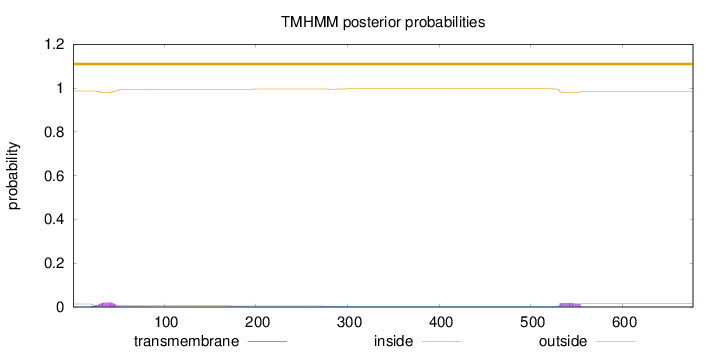
Topology
Subcellular location
Nucleus
Length:
677
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.893429999999999
Exp number, first 60 AAs:
0.42074
Total prob of N-in:
0.01364
outside
1 - 677
Population Genetic Test Statistics
Pi
231.498737
Theta
168.726521
Tajima's D
1.019208
CLR
0.126887
CSRT
0.667666616669166
Interpretation
Uncertain
