Gene
KWMTBOMO10530
Pre Gene Modal
BGIBMGA007133
Annotation
PREDICTED:_TATA-binding_protein-associated_factor_172_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.638 Nuclear Reliability : 1.638
Sequence
CDS
ATGGAGAGCATTAAGAAGGAATGGTGTGAGGAACTTCAGGCGAATTCGGCGAAAACACTGGCGACGCTTTTGGCGCAGCTCGCAGACCGGGACCCCTGCCCTAATAATAAAGTGCTGATTAATTTGAAGGCCTTTCTCAGATGCGACCCGGAGGTGACGCCGCGTGTGTCCTTGGAGCGCGTAGAGGAGGGCGGCGAGGACGGCAGCGGGGACTCCGCCGGGGAGGGACGCCACACGCGCCCTCTCGGACCGCACTTGGACAAGTACAACGGCATCCTGACGCTGCGGGAGCTCCAGCGCAGCGCCGACCGCGCCGCCCCCCGCCGCGGCCGCCCCCCCGCCGCCGCGCACCTCGACCTGGACTCGCTGCTCGACAACGAGGACGAGGCTAGAAAGCTACTTAGAATACAAAGACGAGGCGCGACATTGGCGCTCGTCAGCCTGACGGAACACTTTGGAGACTCACTGCCGCAGAAACTACCCAAGCTGTGGGAGTTTATCACACAACCTTTCGAAAGGGTCCTGACTGACGCCGAATTGGAGAGCATGGACGTGGAGGGTGCCGAGGAGCTGATCTCCCGGCTGCAGGTGTTCGAGGCGGTGTGCGGGCGCGCGGGGGGCGGCGTGCGGGCGGCGCGCGCGGCCGCCGTGTGCTGCGGGTACACGCGCGCGCCCTACACCGCGCTGCGGCACATGGCGGCGCGCGCACTCGCCGCGCTCGCACGCACCGCGCACCCGCACGTCATGCACGCCATCATCGATCAGGTATATTGTAAATGCGTTGTGTAA
Protein
MESIKKEWCEELQANSAKTLATLLAQLADRDPCPNNKVLINLKAFLRCDPEVTPRVSLERVEEGGEDGSGDSAGEGRHTRPLGPHLDKYNGILTLRELQRSADRAAPRRGRPPAAAHLDLDSLLDNEDEARKLLRIQRRGATLALVSLTEHFGDSLPQKLPKLWEFITQPFERVLTDAELESMDVEGAEELISRLQVFEAVCGRAGGGVRAARAAAVCCGYTRAPYTALRHMAARALAALARTAHPHVMHAIIDQVYCKCVV
Summary
Uniprot
H9JC88
A0A0N0PC13
A0A194PII4
A0A2H1VKQ2
A0A212ERX3
A0A2A4JRU2
+ More
A0A3S2LVQ6 D6WW10 A0A1B6GNR9 V5I965 V5GXE7 A0A1B6MAC8 A0A1Y1K022 A0A1Y1K030 A0A067QSZ1 A0A1W4XGH1 A0A2P8XQ60 N6T163 A0A0A9X8J8 W4VRR5 A0A2B4S2D2 A7SE43 A0A224X9F8 T1HUJ9 T1KBT3 A0A1L8DFY6 A0A1L8DH30 A0A023F3Q0 A0A210PRC8 A0A0P5HFR8 A0A0P5IPU3 A0A0P5R2C6 A0A0P6DVL8 A0A0P5R210 A0A0P5HG23 A0A0P5NY89 A0A0P5MJ26 A0A1W7R9G5 A0A0P6DLX6 A0A0P5LHN4 A0A0P5HDC5 A0A0P5QA39 A0A0N8B6D5 A0A0N8BKL4 A0A0P5H150 A0A0P5KLR5 A0A0P5GV70 A0A0P5NH32 A0A0P5LIR3 A0A0P6GN52 A0A0P6EA79 A0A0P5MI20 A0A0N8BMP5 A0A0P5I7V9 A0A0P5GFX7 A0A0P6ELP2 A0A0P5H8L7 A0A0P5QKT2 A0A0P5IVR8 A0A0P5MKA5 A0A0P5GFY6 A0A0P6EEM9 A0A0P6DFS0 E9G250 E2BIA7 A0A250Y891 A0A232EK91 K7JA91 A0A1B6CL15 A0A2Y9RVR9 A0A1S3HG25 A0A1B6DFE2 A0A2Y9D8P3 A0A1S3HME5 G3TLC7 A0A146X650 Q52KH4 A0A384AK17 A0A384AJP4 A0A1B6C462 A0A146P1Z5 A0A3Q0E7G7 A0A1U7TK09 F7DH73 A0A1S2ZBK1 A0A3B4FAW4 K9IQH6 A0A3P4MW44 A0A147AB07 A0A2Y9SMQ4 A0A340YD53 A0A3R7M072 A0A2Y9SKZ0 A0A146X6M8 F1N507 I3MC54 A0A2Y9J2Z5 I3LTQ1 U6DCU6
A0A3S2LVQ6 D6WW10 A0A1B6GNR9 V5I965 V5GXE7 A0A1B6MAC8 A0A1Y1K022 A0A1Y1K030 A0A067QSZ1 A0A1W4XGH1 A0A2P8XQ60 N6T163 A0A0A9X8J8 W4VRR5 A0A2B4S2D2 A7SE43 A0A224X9F8 T1HUJ9 T1KBT3 A0A1L8DFY6 A0A1L8DH30 A0A023F3Q0 A0A210PRC8 A0A0P5HFR8 A0A0P5IPU3 A0A0P5R2C6 A0A0P6DVL8 A0A0P5R210 A0A0P5HG23 A0A0P5NY89 A0A0P5MJ26 A0A1W7R9G5 A0A0P6DLX6 A0A0P5LHN4 A0A0P5HDC5 A0A0P5QA39 A0A0N8B6D5 A0A0N8BKL4 A0A0P5H150 A0A0P5KLR5 A0A0P5GV70 A0A0P5NH32 A0A0P5LIR3 A0A0P6GN52 A0A0P6EA79 A0A0P5MI20 A0A0N8BMP5 A0A0P5I7V9 A0A0P5GFX7 A0A0P6ELP2 A0A0P5H8L7 A0A0P5QKT2 A0A0P5IVR8 A0A0P5MKA5 A0A0P5GFY6 A0A0P6EEM9 A0A0P6DFS0 E9G250 E2BIA7 A0A250Y891 A0A232EK91 K7JA91 A0A1B6CL15 A0A2Y9RVR9 A0A1S3HG25 A0A1B6DFE2 A0A2Y9D8P3 A0A1S3HME5 G3TLC7 A0A146X650 Q52KH4 A0A384AK17 A0A384AJP4 A0A1B6C462 A0A146P1Z5 A0A3Q0E7G7 A0A1U7TK09 F7DH73 A0A1S2ZBK1 A0A3B4FAW4 K9IQH6 A0A3P4MW44 A0A147AB07 A0A2Y9SMQ4 A0A340YD53 A0A3R7M072 A0A2Y9SKZ0 A0A146X6M8 F1N507 I3MC54 A0A2Y9J2Z5 I3LTQ1 U6DCU6
Pubmed
EMBL
BABH01015068
BABH01015069
BABH01015070
BABH01015071
BABH01015072
BABH01015073
+ More
BABH01015074 KQ460711 KPJ12690.1 KQ459603 KPI93127.1 ODYU01003086 SOQ41398.1 AGBW02012899 OWR44242.1 NWSH01000689 PCG74757.1 RSAL01000162 RVE45378.1 KQ971361 EFA08205.1 GECZ01005708 JAS64061.1 GALX01003475 JAB64991.1 GALX01003473 JAB64993.1 GEBQ01007095 JAT32882.1 GEZM01096474 JAV54872.1 GEZM01096473 JAV54874.1 KK852980 KDR13008.1 PYGN01001559 PSN34064.1 APGK01057067 KB741277 ENN71273.1 GBHO01026557 GBRD01014888 JAG17047.1 JAG50938.1 GANO01000305 JAB59566.1 LSMT01000215 PFX23203.1 DS469634 EDO38021.1 GFTR01008892 JAW07534.1 ACPB03015326 CAEY01001955 GFDF01008716 JAV05368.1 GFDF01008322 JAV05762.1 GBBI01003193 JAC15519.1 NEDP02005549 OWF39002.1 GDIQ01228573 JAK23152.1 GDIQ01212082 JAK39643.1 GDIQ01126562 JAL25164.1 GDIQ01075528 JAN19209.1 GDIQ01106616 JAL45110.1 GDIQ01234078 GDIQ01130261 GDIQ01094969 JAK17647.1 GDIQ01136113 JAL15613.1 GDIQ01155896 JAK95829.1 GFAH01000601 JAV47788.1 GDIQ01075527 JAN19210.1 GDIQ01169445 JAK82280.1 GDIQ01230824 GDIQ01228572 JAK23153.1 GDIQ01138658 JAL13068.1 GDIQ01224673 GDIQ01220578 GDIQ01208379 GDIQ01206828 GDIQ01201933 GDIQ01181801 GDIQ01181799 GDIQ01180493 GDIQ01180491 GDIQ01177544 GDIQ01177543 GDIQ01175686 GDIQ01115072 GDIQ01115070 GDIQ01099678 GDIQ01084576 GDIQ01084573 GDIQ01081576 GDIQ01081573 GDIQ01065637 GDIQ01065635 GDIQ01060791 GDIQ01060790 GDIQ01052240 GDIQ01034242 GDIQ01029848 JAK43346.1 GDIQ01223475 GDIQ01222025 GDIQ01178989 GDIQ01178987 GDIQ01168547 GDIQ01165596 GDIQ01161406 GDIQ01098421 GDIQ01067089 GDIQ01067087 GDIQ01033398 JAK83178.1 GDIQ01249178 GDIQ01242971 GDIQ01238726 GDIQ01235229 GDIQ01234077 GDIQ01219379 GDIQ01219377 GDIQ01200734 GDIQ01199440 GDIQ01198187 GDIQ01142559 GDIQ01141406 GDIQ01130668 GDIQ01130263 GDIQ01126561 GDIQ01117568 GDIQ01116268 GDIQ01116266 GDIQ01113818 GDIQ01113816 GDIQ01094968 GDIQ01080279 GDIQ01080276 GDIQ01064234 GDIQ01064232 GDIQ01050755 GDIQ01049342 GDIQ01046199 JAK17648.1 GDIQ01226976 GDIQ01226975 GDIQ01226973 GDIQ01188936 GDIQ01187489 GDIQ01185892 GDIQ01184142 GDIQ01162700 GDIQ01155897 GDIQ01120116 GDIQ01068482 GDIQ01031200 JAK64236.1 GDIQ01237229 JAK14496.1 GDIQ01142557 GDIQ01141407 GDIQ01080277 GDIQ01064231 JAL09169.1 GDIQ01170397 JAK81328.1 GDIQ01032102 JAN62635.1 GDIQ01178986 GDIQ01067086 JAN27651.1 GDIQ01223474 GDIQ01222026 GDIQ01178988 GDIQ01161405 GDIQ01159846 GDIQ01158049 GDIQ01098422 GDIQ01067088 JAK72737.1 GDIQ01226974 GDIQ01188935 GDIQ01187488 GDIQ01185891 GDIQ01184143 GDIQ01184141 GDIQ01162699 GDIQ01068481 JAK89026.1 GDIQ01240473 JAK11252.1 GDIQ01249177 GDIQ01242970 GDIQ01238779 GDIQ01238725 GDIQ01235228 GDIQ01234076 GDIQ01232772 GDIQ01219378 GDIQ01200733 GDIQ01199441 GDIQ01198188 GDIQ01142558 GDIQ01130669 GDIQ01130262 GDIQ01126560 GDIQ01124811 GDIQ01117569 GDIQ01117567 GDIQ01116267 GDIQ01113817 GDIQ01080278 GDIQ01064233 GDIQ01050756 GDIQ01050754 GDIQ01049343 GDIQ01049341 GDIQ01046200 GDIQ01046198 GDIQ01044194 GDIQ01044193 GDIQ01038942 JAK08755.1 GDIQ01181798 GDIQ01180490 GDIQ01177542 GDIQ01084574 GDIQ01081574 GDIQ01065634 GDIQ01060789 JAN33948.1 GDIQ01253432 GDIQ01253430 GDIQ01253429 JAJ98295.1 GDIQ01122714 JAL29012.1 GDIQ01224674 GDIQ01220579 GDIQ01220577 GDIQ01208378 GDIQ01206829 GDIQ01201934 GDIQ01181800 GDIQ01180492 GDIQ01115071 GDIQ01099679 GDIQ01084575 GDIQ01081575 GDIQ01065636 GDIQ01063238 GDIQ01052241 JAK43347.1 GDIQ01158048 JAK93677.1 GDIQ01253431 GDIQ01245171 GDIQ01245170 JAK06555.1 GDIQ01078029 JAN16708.1 GDIQ01078031 GDIQ01078030 JAN16706.1 GL732530 EFX86185.1 GL448480 EFN84571.1 GFFV01000176 JAV39769.1 NNAY01003867 OXU18742.1 AAZX01000485 GEDC01023186 GEDC01014621 JAS14112.1 JAS22677.1 GEDC01012899 JAS24399.1 GCES01049309 JAR37014.1 BC094345 AAH94345.1 GEDC01029006 JAS08292.1 GCES01148353 JAQ37969.1 GABZ01003493 JAA50032.1 CYRY02014205 VCW84410.1 GCES01010607 JAR75716.1 QCYY01002723 ROT68053.1 GCES01049308 JAR37015.1 AGTP01072993 AGTP01072994 AEMK02000096 DQIR01289215 HDC44693.1 HAAF01008597 CCP80421.1
BABH01015074 KQ460711 KPJ12690.1 KQ459603 KPI93127.1 ODYU01003086 SOQ41398.1 AGBW02012899 OWR44242.1 NWSH01000689 PCG74757.1 RSAL01000162 RVE45378.1 KQ971361 EFA08205.1 GECZ01005708 JAS64061.1 GALX01003475 JAB64991.1 GALX01003473 JAB64993.1 GEBQ01007095 JAT32882.1 GEZM01096474 JAV54872.1 GEZM01096473 JAV54874.1 KK852980 KDR13008.1 PYGN01001559 PSN34064.1 APGK01057067 KB741277 ENN71273.1 GBHO01026557 GBRD01014888 JAG17047.1 JAG50938.1 GANO01000305 JAB59566.1 LSMT01000215 PFX23203.1 DS469634 EDO38021.1 GFTR01008892 JAW07534.1 ACPB03015326 CAEY01001955 GFDF01008716 JAV05368.1 GFDF01008322 JAV05762.1 GBBI01003193 JAC15519.1 NEDP02005549 OWF39002.1 GDIQ01228573 JAK23152.1 GDIQ01212082 JAK39643.1 GDIQ01126562 JAL25164.1 GDIQ01075528 JAN19209.1 GDIQ01106616 JAL45110.1 GDIQ01234078 GDIQ01130261 GDIQ01094969 JAK17647.1 GDIQ01136113 JAL15613.1 GDIQ01155896 JAK95829.1 GFAH01000601 JAV47788.1 GDIQ01075527 JAN19210.1 GDIQ01169445 JAK82280.1 GDIQ01230824 GDIQ01228572 JAK23153.1 GDIQ01138658 JAL13068.1 GDIQ01224673 GDIQ01220578 GDIQ01208379 GDIQ01206828 GDIQ01201933 GDIQ01181801 GDIQ01181799 GDIQ01180493 GDIQ01180491 GDIQ01177544 GDIQ01177543 GDIQ01175686 GDIQ01115072 GDIQ01115070 GDIQ01099678 GDIQ01084576 GDIQ01084573 GDIQ01081576 GDIQ01081573 GDIQ01065637 GDIQ01065635 GDIQ01060791 GDIQ01060790 GDIQ01052240 GDIQ01034242 GDIQ01029848 JAK43346.1 GDIQ01223475 GDIQ01222025 GDIQ01178989 GDIQ01178987 GDIQ01168547 GDIQ01165596 GDIQ01161406 GDIQ01098421 GDIQ01067089 GDIQ01067087 GDIQ01033398 JAK83178.1 GDIQ01249178 GDIQ01242971 GDIQ01238726 GDIQ01235229 GDIQ01234077 GDIQ01219379 GDIQ01219377 GDIQ01200734 GDIQ01199440 GDIQ01198187 GDIQ01142559 GDIQ01141406 GDIQ01130668 GDIQ01130263 GDIQ01126561 GDIQ01117568 GDIQ01116268 GDIQ01116266 GDIQ01113818 GDIQ01113816 GDIQ01094968 GDIQ01080279 GDIQ01080276 GDIQ01064234 GDIQ01064232 GDIQ01050755 GDIQ01049342 GDIQ01046199 JAK17648.1 GDIQ01226976 GDIQ01226975 GDIQ01226973 GDIQ01188936 GDIQ01187489 GDIQ01185892 GDIQ01184142 GDIQ01162700 GDIQ01155897 GDIQ01120116 GDIQ01068482 GDIQ01031200 JAK64236.1 GDIQ01237229 JAK14496.1 GDIQ01142557 GDIQ01141407 GDIQ01080277 GDIQ01064231 JAL09169.1 GDIQ01170397 JAK81328.1 GDIQ01032102 JAN62635.1 GDIQ01178986 GDIQ01067086 JAN27651.1 GDIQ01223474 GDIQ01222026 GDIQ01178988 GDIQ01161405 GDIQ01159846 GDIQ01158049 GDIQ01098422 GDIQ01067088 JAK72737.1 GDIQ01226974 GDIQ01188935 GDIQ01187488 GDIQ01185891 GDIQ01184143 GDIQ01184141 GDIQ01162699 GDIQ01068481 JAK89026.1 GDIQ01240473 JAK11252.1 GDIQ01249177 GDIQ01242970 GDIQ01238779 GDIQ01238725 GDIQ01235228 GDIQ01234076 GDIQ01232772 GDIQ01219378 GDIQ01200733 GDIQ01199441 GDIQ01198188 GDIQ01142558 GDIQ01130669 GDIQ01130262 GDIQ01126560 GDIQ01124811 GDIQ01117569 GDIQ01117567 GDIQ01116267 GDIQ01113817 GDIQ01080278 GDIQ01064233 GDIQ01050756 GDIQ01050754 GDIQ01049343 GDIQ01049341 GDIQ01046200 GDIQ01046198 GDIQ01044194 GDIQ01044193 GDIQ01038942 JAK08755.1 GDIQ01181798 GDIQ01180490 GDIQ01177542 GDIQ01084574 GDIQ01081574 GDIQ01065634 GDIQ01060789 JAN33948.1 GDIQ01253432 GDIQ01253430 GDIQ01253429 JAJ98295.1 GDIQ01122714 JAL29012.1 GDIQ01224674 GDIQ01220579 GDIQ01220577 GDIQ01208378 GDIQ01206829 GDIQ01201934 GDIQ01181800 GDIQ01180492 GDIQ01115071 GDIQ01099679 GDIQ01084575 GDIQ01081575 GDIQ01065636 GDIQ01063238 GDIQ01052241 JAK43347.1 GDIQ01158048 JAK93677.1 GDIQ01253431 GDIQ01245171 GDIQ01245170 JAK06555.1 GDIQ01078029 JAN16708.1 GDIQ01078031 GDIQ01078030 JAN16706.1 GL732530 EFX86185.1 GL448480 EFN84571.1 GFFV01000176 JAV39769.1 NNAY01003867 OXU18742.1 AAZX01000485 GEDC01023186 GEDC01014621 JAS14112.1 JAS22677.1 GEDC01012899 JAS24399.1 GCES01049309 JAR37014.1 BC094345 AAH94345.1 GEDC01029006 JAS08292.1 GCES01148353 JAQ37969.1 GABZ01003493 JAA50032.1 CYRY02014205 VCW84410.1 GCES01010607 JAR75716.1 QCYY01002723 ROT68053.1 GCES01049308 JAR37015.1 AGTP01072993 AGTP01072994 AEMK02000096 DQIR01289215 HDC44693.1 HAAF01008597 CCP80421.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000218220
UP000283053
+ More
UP000007266 UP000027135 UP000192223 UP000245037 UP000019118 UP000225706 UP000001593 UP000015103 UP000015104 UP000242188 UP000000305 UP000008237 UP000215335 UP000002358 UP000248480 UP000085678 UP000007646 UP000261681 UP000189704 UP000002280 UP000079721 UP000261460 UP000248484 UP000265300 UP000283509 UP000009136 UP000005215 UP000248482 UP000008227
UP000007266 UP000027135 UP000192223 UP000245037 UP000019118 UP000225706 UP000001593 UP000015103 UP000015104 UP000242188 UP000000305 UP000008237 UP000215335 UP000002358 UP000248480 UP000085678 UP000007646 UP000261681 UP000189704 UP000002280 UP000079721 UP000261460 UP000248484 UP000265300 UP000283509 UP000009136 UP000005215 UP000248482 UP000008227
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JC88
A0A0N0PC13
A0A194PII4
A0A2H1VKQ2
A0A212ERX3
A0A2A4JRU2
+ More
A0A3S2LVQ6 D6WW10 A0A1B6GNR9 V5I965 V5GXE7 A0A1B6MAC8 A0A1Y1K022 A0A1Y1K030 A0A067QSZ1 A0A1W4XGH1 A0A2P8XQ60 N6T163 A0A0A9X8J8 W4VRR5 A0A2B4S2D2 A7SE43 A0A224X9F8 T1HUJ9 T1KBT3 A0A1L8DFY6 A0A1L8DH30 A0A023F3Q0 A0A210PRC8 A0A0P5HFR8 A0A0P5IPU3 A0A0P5R2C6 A0A0P6DVL8 A0A0P5R210 A0A0P5HG23 A0A0P5NY89 A0A0P5MJ26 A0A1W7R9G5 A0A0P6DLX6 A0A0P5LHN4 A0A0P5HDC5 A0A0P5QA39 A0A0N8B6D5 A0A0N8BKL4 A0A0P5H150 A0A0P5KLR5 A0A0P5GV70 A0A0P5NH32 A0A0P5LIR3 A0A0P6GN52 A0A0P6EA79 A0A0P5MI20 A0A0N8BMP5 A0A0P5I7V9 A0A0P5GFX7 A0A0P6ELP2 A0A0P5H8L7 A0A0P5QKT2 A0A0P5IVR8 A0A0P5MKA5 A0A0P5GFY6 A0A0P6EEM9 A0A0P6DFS0 E9G250 E2BIA7 A0A250Y891 A0A232EK91 K7JA91 A0A1B6CL15 A0A2Y9RVR9 A0A1S3HG25 A0A1B6DFE2 A0A2Y9D8P3 A0A1S3HME5 G3TLC7 A0A146X650 Q52KH4 A0A384AK17 A0A384AJP4 A0A1B6C462 A0A146P1Z5 A0A3Q0E7G7 A0A1U7TK09 F7DH73 A0A1S2ZBK1 A0A3B4FAW4 K9IQH6 A0A3P4MW44 A0A147AB07 A0A2Y9SMQ4 A0A340YD53 A0A3R7M072 A0A2Y9SKZ0 A0A146X6M8 F1N507 I3MC54 A0A2Y9J2Z5 I3LTQ1 U6DCU6
A0A3S2LVQ6 D6WW10 A0A1B6GNR9 V5I965 V5GXE7 A0A1B6MAC8 A0A1Y1K022 A0A1Y1K030 A0A067QSZ1 A0A1W4XGH1 A0A2P8XQ60 N6T163 A0A0A9X8J8 W4VRR5 A0A2B4S2D2 A7SE43 A0A224X9F8 T1HUJ9 T1KBT3 A0A1L8DFY6 A0A1L8DH30 A0A023F3Q0 A0A210PRC8 A0A0P5HFR8 A0A0P5IPU3 A0A0P5R2C6 A0A0P6DVL8 A0A0P5R210 A0A0P5HG23 A0A0P5NY89 A0A0P5MJ26 A0A1W7R9G5 A0A0P6DLX6 A0A0P5LHN4 A0A0P5HDC5 A0A0P5QA39 A0A0N8B6D5 A0A0N8BKL4 A0A0P5H150 A0A0P5KLR5 A0A0P5GV70 A0A0P5NH32 A0A0P5LIR3 A0A0P6GN52 A0A0P6EA79 A0A0P5MI20 A0A0N8BMP5 A0A0P5I7V9 A0A0P5GFX7 A0A0P6ELP2 A0A0P5H8L7 A0A0P5QKT2 A0A0P5IVR8 A0A0P5MKA5 A0A0P5GFY6 A0A0P6EEM9 A0A0P6DFS0 E9G250 E2BIA7 A0A250Y891 A0A232EK91 K7JA91 A0A1B6CL15 A0A2Y9RVR9 A0A1S3HG25 A0A1B6DFE2 A0A2Y9D8P3 A0A1S3HME5 G3TLC7 A0A146X650 Q52KH4 A0A384AK17 A0A384AJP4 A0A1B6C462 A0A146P1Z5 A0A3Q0E7G7 A0A1U7TK09 F7DH73 A0A1S2ZBK1 A0A3B4FAW4 K9IQH6 A0A3P4MW44 A0A147AB07 A0A2Y9SMQ4 A0A340YD53 A0A3R7M072 A0A2Y9SKZ0 A0A146X6M8 F1N507 I3MC54 A0A2Y9J2Z5 I3LTQ1 U6DCU6
PDB
6G7E
E-value=0.00622898,
Score=91
Ontologies
GO
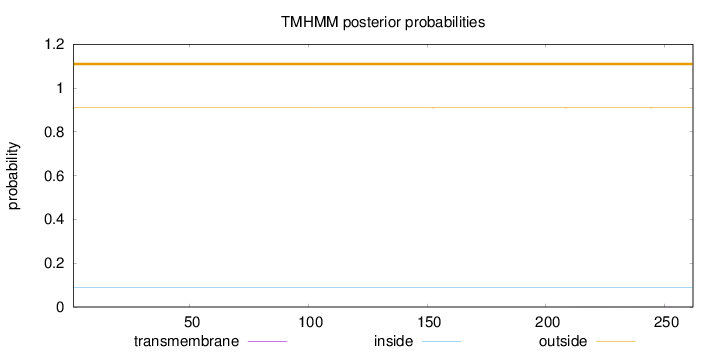
Topology
Length:
262
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01154
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.09037
outside
1 - 262
Population Genetic Test Statistics
Pi
189.292892
Theta
176.090145
Tajima's D
-1.440146
CLR
104.039076
CSRT
0.0694465276736163
Interpretation
Uncertain
