Gene
KWMTBOMO10529
Pre Gene Modal
BGIBMGA007133
Annotation
PREDICTED:_TATA-binding_protein-associated_factor_172_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.827
Sequence
CDS
ATGGCTACAAGAAGTGTCATAATAGGCCTCTACAAATTCCCGATCGATTTGGCAGACGTGCTACCTCGTCTGTGGCCGTATTTAGACCATGCGACCTCATCAGTGCGGAAGGCCACACTACAGACTCTCCGCACCCTGACGCGGCCCCTGGTCACCACAGCCGCACCGCGCGCCGACAGCGCCCACAGCCCGCCCCCTGACAACGCCGCGCCCAGTGAAGCCCCACCGAACGGGCTGGACGAACAGTACTTGATGTGGACCCCGGAGCTGTTGCAGGAAGCAATGAGACACGTTTACCAGAGAGTGTTGTTCGAACATATCTATGAAATACAAGAAATTGCAGTGCAAGTGTGGGAGAACCTGATCCGGCACGCGTCCCTGGGCGTGATCCTGGTGGCGGCGTGTCCCATGCTGTCCACGTGGCTGTGCCTGGCCATGCAGCCCGCGCGCCTGGCCGTGGACCCGGCGCTCATACTCGTGCCGCCGCTCAAGGAGCGGCGCACGCGCACGAGCCCGGCGGAGGCGGTGCCGGGCGAGGTGCGCGCGGCGCTGCGCTGGTACGTGGGCGGCGCCGACACGCAGGCCAGCGCCGCGCGGGACAAGAACGTGTCGCGGGCGCGCTGTCTCGCCGCCGACATACTGGGGCACCTGTCGTGCTACCTGGTGCAGCCCGCGCCCGGCATCGACTACACGGCGGAAGAAGAGAGCCCCATCGACTGTTACGTGAAGGTGATGGTGGTGTACCTCCGCTCGGGCAGCGCCCTGCAGCGCCTGGTGGCCAGCCTCGTAGTGTCGGCCTGGGCGCGCCACACGCGGCGCCGCGGCCTCTACCCGCCCCCCGCCCCGCGCCCCGCACACCCGGATGAAGAGAAAGATGAAAATTATGAAACGAACGACATATCAAAGCTGGCTCCGGCGTCGCTGGTGTCCACCCTGCACACGGCGCTCAACCAGGCGCTGTACTATGACGAGGTGGCTCTCAACTGCAACAGGATACTCCAAGAGGCAAGAGATTTTTTGGCGACCTTGAAACATTACAAGTTACCAGTTGACGGTGAGGAGTTCAACAATATTGTGAGATTGGAACAGGTGGTGCAGCTGACGGGCGCCGCGCAGGGCATGGTGAGCCGCATGAAGCCCAAGCGAGCCGCGCCCGCGCTGGACGAGCGCCGCCGCGCGCTGCGGGCCGCCGCCGCGCACACGCACGCCATGCACGCCGCGCTCGCCGTCGCGTACGTACCCCGCCACCGCAGATCAACACATTACACTTGA
Protein
MATRSVIIGLYKFPIDLADVLPRLWPYLDHATSSVRKATLQTLRTLTRPLVTTAAPRADSAHSPPPDNAAPSEAPPNGLDEQYLMWTPELLQEAMRHVYQRVLFEHIYEIQEIAVQVWENLIRHASLGVILVAACPMLSTWLCLAMQPARLAVDPALILVPPLKERRTRTSPAEAVPGEVRAALRWYVGGADTQASAARDKNVSRARCLAADILGHLSCYLVQPAPGIDYTAEEESPIDCYVKVMVVYLRSGSALQRLVASLVVSAWARHTRRRGLYPPPAPRPAHPDEEKDENYETNDISKLAPASLVSTLHTALNQALYYDEVALNCNRILQEARDFLATLKHYKLPVDGEEFNNIVRLEQVVQLTGAAQGMVSRMKPKRAAPALDERRRALRAAAAHTHAMHAALAVAYVPRHRRSTHYT
Summary
Uniprot
H9JC88
A0A2H1VKQ2
A0A194PII4
A0A0N0PC13
A0A212ERX3
A0A067QSZ1
+ More
A0A1B6DFE2 A0A1B6CL15 A0A1B6C462 A0A1B6MAC8 A0A2P8XQ60 V5I965 A0A1Y1K030 A0A1Y1K022 V5GXE7 A0A1W4XGH1 A0A336KBP6 A0A182N4Z9 A0A0T6B0J0 W4VRR5 A0A182P5N4 A0A182VQT4 A0A182JX29 A0A182M311 A0A182IRV5 D6WW10 B4QXR9 J9XXX0 Q9VF02 A0A182RP11 B4HLF4 A0A084VRP0 A0A182F6W2 A0A182XVA4 B4PR56 A0A1Q3FBY8 A0A1Q3FES8 B3M3A2 Q71V44 A0A182G5L0 A0A1B0GDV1 A0A182SC65 B0W966 A0A1A9V2J1 B3P3Y2 A0A1B0C5A7 Q16JW5 U4TV80 A0A1B0AJ89 A0A182X2P4 A0A182QMB3 A0A0L0C794 N6T163 A0A182UMS2 A0A2M4A3A0 A0A2M4A2S2 A0A1A9X433 A0A182HGR5 Q7PUQ9 A0A0M9A581 A0A310SVY7 W5JFS1 B4N990 A0A2M4CNY7 B4K5P5 Q296N6 A0A1W4VQF4 A0A1I8PPP2 A0A0M4EUQ3 B4GEW7 A0A0K8V4N8 A0A088A8J0 K7JA91 A0A2A3E2Q1 A0A034VBP8 A0A0L7R0W2 A0A232EK91 A0A3B0KDI2 A0A1L8DFY6 B4JT60 A0A1L8DH30 B4LXA1 A0A1J1HY29 E2BIA7 A0A023F3Q0 W8BMY6 T1HUJ9 E0VSS7 A0A224X9F8 A0A026WRV5 A0A1I8MMC4 A0A0A9X8J8 T1SF24 A0A195EV94 A0A0P5YDY7 A0A0P4YCL3 A0A0P5XI11 A0A0P4XWI0
A0A1B6DFE2 A0A1B6CL15 A0A1B6C462 A0A1B6MAC8 A0A2P8XQ60 V5I965 A0A1Y1K030 A0A1Y1K022 V5GXE7 A0A1W4XGH1 A0A336KBP6 A0A182N4Z9 A0A0T6B0J0 W4VRR5 A0A182P5N4 A0A182VQT4 A0A182JX29 A0A182M311 A0A182IRV5 D6WW10 B4QXR9 J9XXX0 Q9VF02 A0A182RP11 B4HLF4 A0A084VRP0 A0A182F6W2 A0A182XVA4 B4PR56 A0A1Q3FBY8 A0A1Q3FES8 B3M3A2 Q71V44 A0A182G5L0 A0A1B0GDV1 A0A182SC65 B0W966 A0A1A9V2J1 B3P3Y2 A0A1B0C5A7 Q16JW5 U4TV80 A0A1B0AJ89 A0A182X2P4 A0A182QMB3 A0A0L0C794 N6T163 A0A182UMS2 A0A2M4A3A0 A0A2M4A2S2 A0A1A9X433 A0A182HGR5 Q7PUQ9 A0A0M9A581 A0A310SVY7 W5JFS1 B4N990 A0A2M4CNY7 B4K5P5 Q296N6 A0A1W4VQF4 A0A1I8PPP2 A0A0M4EUQ3 B4GEW7 A0A0K8V4N8 A0A088A8J0 K7JA91 A0A2A3E2Q1 A0A034VBP8 A0A0L7R0W2 A0A232EK91 A0A3B0KDI2 A0A1L8DFY6 B4JT60 A0A1L8DH30 B4LXA1 A0A1J1HY29 E2BIA7 A0A023F3Q0 W8BMY6 T1HUJ9 E0VSS7 A0A224X9F8 A0A026WRV5 A0A1I8MMC4 A0A0A9X8J8 T1SF24 A0A195EV94 A0A0P5YDY7 A0A0P4YCL3 A0A0P5XI11 A0A0P4XWI0
Pubmed
19121390
26354079
22118469
24845553
29403074
28004739
+ More
18362917 19820115 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24438588 25244985 17550304 8774890 26483478 17510324 23537049 26108605 12364791 14747013 17210077 20920257 23761445 15632085 20075255 25348373 28648823 20798317 25474469 24495485 20566863 24508170 25315136 25401762
18362917 19820115 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24438588 25244985 17550304 8774890 26483478 17510324 23537049 26108605 12364791 14747013 17210077 20920257 23761445 15632085 20075255 25348373 28648823 20798317 25474469 24495485 20566863 24508170 25315136 25401762
EMBL
BABH01015068
BABH01015069
BABH01015070
BABH01015071
BABH01015072
BABH01015073
+ More
BABH01015074 ODYU01003086 SOQ41398.1 KQ459603 KPI93127.1 KQ460711 KPJ12690.1 AGBW02012899 OWR44242.1 KK852980 KDR13008.1 GEDC01012899 JAS24399.1 GEDC01023186 GEDC01014621 JAS14112.1 JAS22677.1 GEDC01029006 JAS08292.1 GEBQ01007095 JAT32882.1 PYGN01001559 PSN34064.1 GALX01003475 JAB64991.1 GEZM01096473 JAV54874.1 GEZM01096474 JAV54872.1 GALX01003473 JAB64993.1 UFQS01000046 UFQT01000046 SSW98500.1 SSX18886.1 LJIG01016429 KRT80629.1 GANO01000305 JAB59566.1 AXCM01001504 KQ971361 EFA08205.1 CM000364 EDX12743.1 BT149864 AFS34653.1 AE014297 AAF55260.3 AHN57350.1 CH480815 EDW41974.1 ATLV01015761 KE525036 KFB40634.1 CM000160 EDW97378.1 GFDL01010023 JAV25022.1 GFDL01008997 JAV26048.1 CH902617 EDV43563.1 AF033104 AAB95091.3 JXUM01043776 JXUM01043777 JXUM01043778 CCAG010012029 DS231862 EDS39763.1 CH954181 EDV49088.1 JXJN01025919 CH477989 EAT34577.1 KB631617 ERL84707.1 AXCN02000181 JRES01000819 KNC28122.1 APGK01057067 KB741277 ENN71273.1 GGFK01001797 MBW35118.1 GGFK01001689 MBW35010.1 APCN01005942 AAAB01008987 EAA01172.6 KQ435732 KOX77487.1 KQ759894 OAD62043.1 ADMH02001524 ETN62193.1 CH964232 EDW81637.1 GGFL01002859 MBW67037.1 CH933806 EDW15107.1 CM000070 EAL28421.2 CP012526 ALC46880.1 CH479182 EDW34152.1 GDHF01018412 JAI33902.1 AAZX01000485 KZ288419 PBC25985.1 GAKP01018256 JAC40696.1 KQ414669 KOC64488.1 NNAY01003867 OXU18742.1 OUUW01000008 SPP84299.1 GFDF01008716 JAV05368.1 CH916373 EDV94950.1 GFDF01008322 JAV05762.1 CH940650 EDW66753.2 CVRI01000036 CRK93000.1 GL448480 EFN84571.1 GBBI01003193 JAC15519.1 GAMC01011820 GAMC01011819 JAB94736.1 ACPB03015326 DS235756 EEB16433.1 GFTR01008892 JAW07534.1 KK107119 EZA58663.1 GBHO01026557 GBRD01014888 JAG17047.1 JAG50938.1 JX984954 AGS58422.1 KQ981954 KYN32163.1 GDIP01061027 JAM42688.1 GDIP01230311 JAI93090.1 GDIP01071929 JAM31786.1 GDIP01238016 JAI85385.1
BABH01015074 ODYU01003086 SOQ41398.1 KQ459603 KPI93127.1 KQ460711 KPJ12690.1 AGBW02012899 OWR44242.1 KK852980 KDR13008.1 GEDC01012899 JAS24399.1 GEDC01023186 GEDC01014621 JAS14112.1 JAS22677.1 GEDC01029006 JAS08292.1 GEBQ01007095 JAT32882.1 PYGN01001559 PSN34064.1 GALX01003475 JAB64991.1 GEZM01096473 JAV54874.1 GEZM01096474 JAV54872.1 GALX01003473 JAB64993.1 UFQS01000046 UFQT01000046 SSW98500.1 SSX18886.1 LJIG01016429 KRT80629.1 GANO01000305 JAB59566.1 AXCM01001504 KQ971361 EFA08205.1 CM000364 EDX12743.1 BT149864 AFS34653.1 AE014297 AAF55260.3 AHN57350.1 CH480815 EDW41974.1 ATLV01015761 KE525036 KFB40634.1 CM000160 EDW97378.1 GFDL01010023 JAV25022.1 GFDL01008997 JAV26048.1 CH902617 EDV43563.1 AF033104 AAB95091.3 JXUM01043776 JXUM01043777 JXUM01043778 CCAG010012029 DS231862 EDS39763.1 CH954181 EDV49088.1 JXJN01025919 CH477989 EAT34577.1 KB631617 ERL84707.1 AXCN02000181 JRES01000819 KNC28122.1 APGK01057067 KB741277 ENN71273.1 GGFK01001797 MBW35118.1 GGFK01001689 MBW35010.1 APCN01005942 AAAB01008987 EAA01172.6 KQ435732 KOX77487.1 KQ759894 OAD62043.1 ADMH02001524 ETN62193.1 CH964232 EDW81637.1 GGFL01002859 MBW67037.1 CH933806 EDW15107.1 CM000070 EAL28421.2 CP012526 ALC46880.1 CH479182 EDW34152.1 GDHF01018412 JAI33902.1 AAZX01000485 KZ288419 PBC25985.1 GAKP01018256 JAC40696.1 KQ414669 KOC64488.1 NNAY01003867 OXU18742.1 OUUW01000008 SPP84299.1 GFDF01008716 JAV05368.1 CH916373 EDV94950.1 GFDF01008322 JAV05762.1 CH940650 EDW66753.2 CVRI01000036 CRK93000.1 GL448480 EFN84571.1 GBBI01003193 JAC15519.1 GAMC01011820 GAMC01011819 JAB94736.1 ACPB03015326 DS235756 EEB16433.1 GFTR01008892 JAW07534.1 KK107119 EZA58663.1 GBHO01026557 GBRD01014888 JAG17047.1 JAG50938.1 JX984954 AGS58422.1 KQ981954 KYN32163.1 GDIP01061027 JAM42688.1 GDIP01230311 JAI93090.1 GDIP01071929 JAM31786.1 GDIP01238016 JAI85385.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000027135
UP000245037
+ More
UP000192223 UP000075884 UP000075885 UP000075920 UP000075881 UP000075883 UP000075880 UP000007266 UP000000304 UP000000803 UP000075900 UP000001292 UP000030765 UP000069272 UP000076408 UP000002282 UP000007801 UP000069940 UP000092444 UP000075901 UP000002320 UP000078200 UP000008711 UP000092460 UP000008820 UP000030742 UP000092445 UP000076407 UP000075886 UP000037069 UP000019118 UP000075903 UP000091820 UP000075840 UP000007062 UP000053105 UP000000673 UP000007798 UP000009192 UP000001819 UP000192221 UP000095300 UP000092553 UP000008744 UP000005203 UP000002358 UP000242457 UP000053825 UP000215335 UP000268350 UP000001070 UP000008792 UP000183832 UP000008237 UP000015103 UP000009046 UP000053097 UP000095301 UP000078541
UP000192223 UP000075884 UP000075885 UP000075920 UP000075881 UP000075883 UP000075880 UP000007266 UP000000304 UP000000803 UP000075900 UP000001292 UP000030765 UP000069272 UP000076408 UP000002282 UP000007801 UP000069940 UP000092444 UP000075901 UP000002320 UP000078200 UP000008711 UP000092460 UP000008820 UP000030742 UP000092445 UP000076407 UP000075886 UP000037069 UP000019118 UP000075903 UP000091820 UP000075840 UP000007062 UP000053105 UP000000673 UP000007798 UP000009192 UP000001819 UP000192221 UP000095300 UP000092553 UP000008744 UP000005203 UP000002358 UP000242457 UP000053825 UP000215335 UP000268350 UP000001070 UP000008792 UP000183832 UP000008237 UP000015103 UP000009046 UP000053097 UP000095301 UP000078541
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JC88
A0A2H1VKQ2
A0A194PII4
A0A0N0PC13
A0A212ERX3
A0A067QSZ1
+ More
A0A1B6DFE2 A0A1B6CL15 A0A1B6C462 A0A1B6MAC8 A0A2P8XQ60 V5I965 A0A1Y1K030 A0A1Y1K022 V5GXE7 A0A1W4XGH1 A0A336KBP6 A0A182N4Z9 A0A0T6B0J0 W4VRR5 A0A182P5N4 A0A182VQT4 A0A182JX29 A0A182M311 A0A182IRV5 D6WW10 B4QXR9 J9XXX0 Q9VF02 A0A182RP11 B4HLF4 A0A084VRP0 A0A182F6W2 A0A182XVA4 B4PR56 A0A1Q3FBY8 A0A1Q3FES8 B3M3A2 Q71V44 A0A182G5L0 A0A1B0GDV1 A0A182SC65 B0W966 A0A1A9V2J1 B3P3Y2 A0A1B0C5A7 Q16JW5 U4TV80 A0A1B0AJ89 A0A182X2P4 A0A182QMB3 A0A0L0C794 N6T163 A0A182UMS2 A0A2M4A3A0 A0A2M4A2S2 A0A1A9X433 A0A182HGR5 Q7PUQ9 A0A0M9A581 A0A310SVY7 W5JFS1 B4N990 A0A2M4CNY7 B4K5P5 Q296N6 A0A1W4VQF4 A0A1I8PPP2 A0A0M4EUQ3 B4GEW7 A0A0K8V4N8 A0A088A8J0 K7JA91 A0A2A3E2Q1 A0A034VBP8 A0A0L7R0W2 A0A232EK91 A0A3B0KDI2 A0A1L8DFY6 B4JT60 A0A1L8DH30 B4LXA1 A0A1J1HY29 E2BIA7 A0A023F3Q0 W8BMY6 T1HUJ9 E0VSS7 A0A224X9F8 A0A026WRV5 A0A1I8MMC4 A0A0A9X8J8 T1SF24 A0A195EV94 A0A0P5YDY7 A0A0P4YCL3 A0A0P5XI11 A0A0P4XWI0
A0A1B6DFE2 A0A1B6CL15 A0A1B6C462 A0A1B6MAC8 A0A2P8XQ60 V5I965 A0A1Y1K030 A0A1Y1K022 V5GXE7 A0A1W4XGH1 A0A336KBP6 A0A182N4Z9 A0A0T6B0J0 W4VRR5 A0A182P5N4 A0A182VQT4 A0A182JX29 A0A182M311 A0A182IRV5 D6WW10 B4QXR9 J9XXX0 Q9VF02 A0A182RP11 B4HLF4 A0A084VRP0 A0A182F6W2 A0A182XVA4 B4PR56 A0A1Q3FBY8 A0A1Q3FES8 B3M3A2 Q71V44 A0A182G5L0 A0A1B0GDV1 A0A182SC65 B0W966 A0A1A9V2J1 B3P3Y2 A0A1B0C5A7 Q16JW5 U4TV80 A0A1B0AJ89 A0A182X2P4 A0A182QMB3 A0A0L0C794 N6T163 A0A182UMS2 A0A2M4A3A0 A0A2M4A2S2 A0A1A9X433 A0A182HGR5 Q7PUQ9 A0A0M9A581 A0A310SVY7 W5JFS1 B4N990 A0A2M4CNY7 B4K5P5 Q296N6 A0A1W4VQF4 A0A1I8PPP2 A0A0M4EUQ3 B4GEW7 A0A0K8V4N8 A0A088A8J0 K7JA91 A0A2A3E2Q1 A0A034VBP8 A0A0L7R0W2 A0A232EK91 A0A3B0KDI2 A0A1L8DFY6 B4JT60 A0A1L8DH30 B4LXA1 A0A1J1HY29 E2BIA7 A0A023F3Q0 W8BMY6 T1HUJ9 E0VSS7 A0A224X9F8 A0A026WRV5 A0A1I8MMC4 A0A0A9X8J8 T1SF24 A0A195EV94 A0A0P5YDY7 A0A0P4YCL3 A0A0P5XI11 A0A0P4XWI0
Ontologies
GO
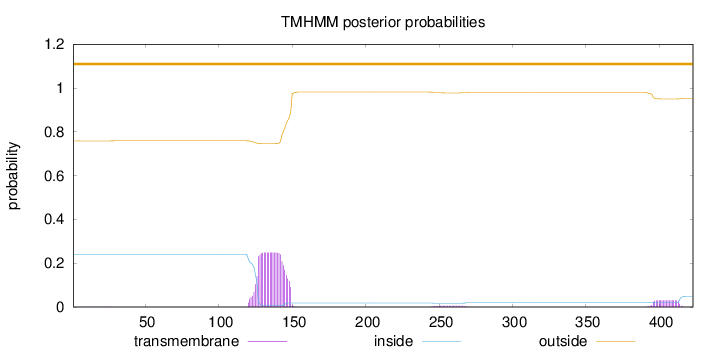
Topology
Length:
423
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
6.1744
Exp number, first 60 AAs:
0.02463
Total prob of N-in:
0.24138
outside
1 - 423
Population Genetic Test Statistics
Pi
145.485196
Theta
132.249367
Tajima's D
0.946454
CLR
0.234202
CSRT
0.641517924103795
Interpretation
Uncertain
