Gene
KWMTBOMO10526
Pre Gene Modal
BGIBMGA007132
Annotation
PREDICTED:_endonuclease_III-like_protein_1_isoform_X3_[Bombyx_mori]
Full name
Endonuclease III homolog
Alternative Name
Bifunctional DNA N-glycosylase/DNA-(apurinic or apyrimidinic site) lyase
Location in the cell
Nuclear Reliability : 4.282
Sequence
CDS
ATGGAGAACAGGAACAAGGTTACAGAAAACAACATTGGCTCAGAGACCGCCATAGCAGTACCTGATTTAAGCCAATTCAAATTCGAGAAAAAATCTCATGTAAAAATTAAATTTGAAAAAGAATCTCCAAGCAAGCAAGAACCTGAAGGCTTATGGGAACCACCAAATTGGAGACAGTTTTTTTTGAACCTGAAGATTATGAGAGCTAATAATGATGCTCCAGTAGATACAATGGGATGTCATATGAACATGGATGAGAATGTACCACCAGAGGTGTACAGATATCAATGCTTAGTTTCTCTGATGCTATCAAGCCAAACCAAGGACCAAGTTAACTTTACTGCTATGCAGAGGCTTCGGGCTCGGGGTCTTACAGTTGATAATGTGTTGAGTATGAGTGATGAAGAACTAGGTCAGTTGATATATCCTGTTGGATTTTGGAAGACAAAAGTAAAATACATAAAAAAAACAACAGGAATTCTAAAAGAGCAGTACAATGGAGACATCCCCGATTCAGTCGAAAAGTTGTGCAAACTGAATGGTGTTGGTCCTAAAATGGCACATATTTGTATGAAGGTTGCTTGGAACAAGATCACTGGAATCGGTGTTGACACACATGTACATAGGATATGCAATAGGATTGGATGGGTCAAAAAACCAACTAAAACTCCTGAGGAAACTCGCCTAGCTCTACAGAGTTGGTTGCCTTTTGAACAATGGAGTGAAGTTAATCATCTACTTGTTGGATTTGGCCAAACTATATGTCAACCAATAAAACCTAATTGTGGAGAATGCCTGAATAATGATATTTGCCCTGCAAGTACTATAGGGAAAAAATCGCCAAAAAAATCTGACAAACAAACCAGAGATAAAATTGAAGTTGGAATTAAAATTGAAGCTGATATGGAAGATTTTCAAGTAAAAAATGAAAAGACTTGGAAAACTATAAGTCCAAAAGTGAATACCAAGATTCAAGATTTAGAATTGGACAAAATTAATACATCTACACCACAAAAACATCAAGATAAATTAAAAAAACCAGAATTCAATAACCAATTTGAATTACATAGTGATGCGACGTCAAATGAAAATAATAGTACAAAAGCTTCCAAGTCCTATGAAAATCTTAAACATACGAAAGACACTTGTGGTGGTAAAAGAATATTCAAAAAAGCAGATGTCCATTGCGATGAGCCTAAATTACCTTTAAAAAGAAAATCGCCTAGAGTTAATCAAAGAACAGAAACTAAAAAACTGGAAAATTTAAAAAAAATTAGAACACGGTCAGCACTAAAATAA
Protein
MENRNKVTENNIGSETAIAVPDLSQFKFEKKSHVKIKFEKESPSKQEPEGLWEPPNWRQFFLNLKIMRANNDAPVDTMGCHMNMDENVPPEVYRYQCLVSLMLSSQTKDQVNFTAMQRLRARGLTVDNVLSMSDEELGQLIYPVGFWKTKVKYIKKTTGILKEQYNGDIPDSVEKLCKLNGVGPKMAHICMKVAWNKITGIGVDTHVHRICNRIGWVKKPTKTPEETRLALQSWLPFEQWSEVNHLLVGFGQTICQPIKPNCGECLNNDICPASTIGKKSPKKSDKQTRDKIEVGIKIEADMEDFQVKNEKTWKTISPKVNTKIQDLELDKINTSTPQKHQDKLKKPEFNNQFELHSDATSNENNSTKASKSYENLKHTKDTCGGKRIFKKADVHCDEPKLPLKRKSPRVNQRTETKKLENLKKIRTRSALK
Summary
Description
Bifunctional DNA N-glycosylase with associated apurinic/apyrimidinic (AP) lyase function that catalyzes the first step in base excision repair (BER), the primary repair pathway for the repair of oxidative DNA damage. The DNA N-glycosylase activity releases the damaged DNA base from DNA by cleaving the N-glycosidic bond, leaving an AP site. The AP lyase activity cleaves the phosphodiester bond 3' to the AP site by a beta-elimination. Primarily recognizes and repairs oxidative base damage of pyrimidines.
Catalytic Activity
The C-O-P bond 3' to the apurinic or apyrimidinic site in DNA is broken by a beta-elimination reaction, leaving a 3'-terminal unsaturated sugar and a product with a terminal 5'-phosphate.
Cofactor
[4Fe-4S] cluster
Similarity
Belongs to the Nth/MutY family.
Uniprot
EC Number
3.2.2.-
4.2.99.18
4.2.99.18
Proteomes
PRIDE
Interpro
SUPFAM
SSF48150
SSF48150
Gene 3D
CDD
ProteinModelPortal
PDB
1ORP
E-value=1.91587e-20,
Score=245
Ontologies
GO
Topology
Subcellular location
Nucleus
Mitochondrion
Mitochondrion
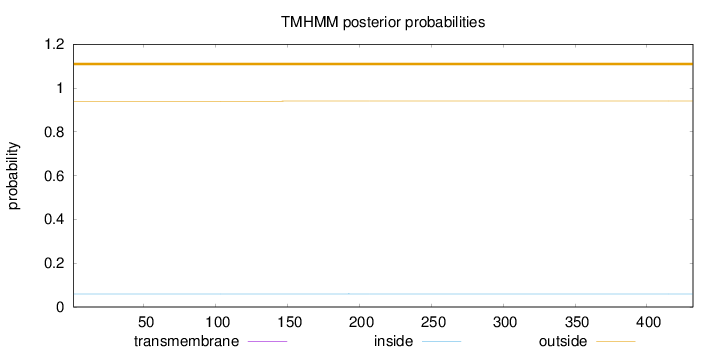
Length:
432
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0023
Exp number, first 60 AAs:
0.00107
Total prob of N-in:
0.05983
outside
1 - 432
Population Genetic Test Statistics
Pi
287.746549
Theta
210.805269
Tajima's D
1.205517
CLR
0.000021
CSRT
0.713364331783411
Interpretation
Uncertain
