Gene
KWMTBOMO10524 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007131
Annotation
PREDICTED:_developmentally-regulated_GTP-binding_protein_2_[Papilio_polytes]
Location in the cell
Cytoplasmic Reliability : 3.265
Sequence
CDS
ATGGGGATTTTAGAGAAAATATCAGAAATTGAGAAAGAAATCGCGAGAACACAGAAAAATAAAGCTACTGAATATCACTTGGGTCTCTTAAAAGCCAAATTAGCAAAATATAGATCACAGTTGCTGGAACCTTCAAAAAAAGGTGGAGACAAAGGTGAAGGTTTCGATGTGCTAAAGTCTGGTGATGCTCGAGTTGCTTTGATAGGCTTTCCTTCTGTGGGTAAGTCGACCCTTTTATCGACCCTCACTCACACACATTCAGAAGCAGCAAGTTATGAATTCACAACGCTGACATGTATACCTGGAGTTATAGAATATCGCGGTGCCAACATACAATTGTTAGATTTGCCTGGAATTATTGAAGGTGCTGCTCAGGGCAAAGGTAGAGGAAGACAGGTTATAGCCGTAGCACGTACTGCAGACTTGGTATTAATGATGCTGGATGCTACAAAACCATATGTGCATAGACAACTTCTAGAAAAAGAATTAGAGAGCGTCGGCATCCGTCTGAATAAACCTAAACCTAATATTTATTTTAAGGTGAAAAAAGGTGGTGGCTTATCATTTAACTCAACTTGTCAACTAACTAAAATTGATGAGAAAATGGTGCAAATGATCTTACATGAATACAAGATATTTAATGCTGAGGTTCTGTTCCGTGAAGACTGCACACCGGACGAGCTCATAGATGTTATACTCGCGAACAGAGTGTACCTGCCCTGCCTCTACGTCTACAACAAGATAGATCAGATCTCCATCCAGGAAGTCGACAGGATAGCGAGACAACCGCATTCTGTAGTCGTTAGTTGTAATATGAAACTGAATTTGGATTATCTGCTAGAGATACTCTGGGAATATCTGTCTCTTATCAGGGTATATACCAAAAAGCCTGGACAGCCCCCAGATTTTGATGATGGACTTATCCTGAGAAAGGGTGTGACCGTGGAGCACGTCTGCCACTCCATCCACCGGACCCTGGCCGCCCAGCTGAAATACGCGCTGGTGTGGGGCACCAGTACCAAGTACTCTCCGCAGCGTGTCGGCATCAACCACACCGTCAACGATGAAGATGTCGTCCAACTTATCAAGAAATAA
Protein
MGILEKISEIEKEIARTQKNKATEYHLGLLKAKLAKYRSQLLEPSKKGGDKGEGFDVLKSGDARVALIGFPSVGKSTLLSTLTHTHSEAASYEFTTLTCIPGVIEYRGANIQLLDLPGIIEGAAQGKGRGRQVIAVARTADLVLMMLDATKPYVHRQLLEKELESVGIRLNKPKPNIYFKVKKGGGLSFNSTCQLTKIDEKMVQMILHEYKIFNAEVLFREDCTPDELIDVILANRVYLPCLYVYNKIDQISIQEVDRIARQPHSVVVSCNMKLNLDYLLEILWEYLSLIRVYTKKPGQPPDFDDGLILRKGVTVEHVCHSIHRTLAAQLKYALVWGTSTKYSPQRVGINHTVNDEDVVQLIKK
Summary
Uniprot
H9JC86
A0A194PJA8
A0A2H1VPR7
A0A0N1IP67
S4P883
A0A212ERW3
+ More
A0A0C9PR51 E2A6X4 A0A232F7C7 K7IZ66 A0A026WHC5 A0A195DB47 F4W717 A0A195AXY8 D6WYF1 E2BZ72 A0A158NM95 A0A151K0R2 A0A151X7X2 A0A0L7R4R3 A0A2A3E297 A0A088AQW7 N6TS78 A0A1Y1M6R3 B4M4Q0 J3JWM1 A0A0M9AAV2 A0A195C2K7 B4NF05 B3LXQ6 B4K4T9 A0A1B6EKJ2 A0A2J7REX1 A0A1W4WRG1 A0A3B0JNL9 A0A0N7Z918 Q293L9 A0A067R6W0 R4G4S6 A0A1W4W8E4 T1PGL0 A0A224XSJ5 B4PNI6 A0A2R7VPY5 A0A0M4F532 B4JT29 A0A0A9Y113 B3P096 B4QSV7 J9JW82 B4IHT4 Q9VDV2 A0A2S2NUG2 A0A1B6MC88 A0A1I8PZ11 W8ATU1 A0A1A9YSC9 A0A1A9VTC3 A0A1B0BPK8 A0A1A9ZPP7 A0A1A9WTX5 D3TP62 A0A0L0BST2 A0A2M4BRW0 A0A2M3Z2K6 A0A2M4A8M5 A0A182FA84 A0A034VDF5 A0A1L8DMK3 T1DQ63 W5J2M0 A0A336M8B8 A0A182RFV3 A0A0K8WF15 A0A182XWJ0 A0A182PD55 A0A182QQB5 A0A182M5T7 A0A1Q3F6F7 A0A182VYD3 A0A182VP02 A0A182K5W5 A0A182WZI1 A0A182LQN9 Q7PTK7 A0A182HZM8 A0A182NMV3 B0X4C9 A0A182U039 E0VST7 A0A182IT76 Q16MM8 A0A310SHU0 A0A023ERT8 A0A1D2MN74 U5EVM9 A0A154PFD8 A0A164UAD1 A0A0P6GWU6 E9G055
A0A0C9PR51 E2A6X4 A0A232F7C7 K7IZ66 A0A026WHC5 A0A195DB47 F4W717 A0A195AXY8 D6WYF1 E2BZ72 A0A158NM95 A0A151K0R2 A0A151X7X2 A0A0L7R4R3 A0A2A3E297 A0A088AQW7 N6TS78 A0A1Y1M6R3 B4M4Q0 J3JWM1 A0A0M9AAV2 A0A195C2K7 B4NF05 B3LXQ6 B4K4T9 A0A1B6EKJ2 A0A2J7REX1 A0A1W4WRG1 A0A3B0JNL9 A0A0N7Z918 Q293L9 A0A067R6W0 R4G4S6 A0A1W4W8E4 T1PGL0 A0A224XSJ5 B4PNI6 A0A2R7VPY5 A0A0M4F532 B4JT29 A0A0A9Y113 B3P096 B4QSV7 J9JW82 B4IHT4 Q9VDV2 A0A2S2NUG2 A0A1B6MC88 A0A1I8PZ11 W8ATU1 A0A1A9YSC9 A0A1A9VTC3 A0A1B0BPK8 A0A1A9ZPP7 A0A1A9WTX5 D3TP62 A0A0L0BST2 A0A2M4BRW0 A0A2M3Z2K6 A0A2M4A8M5 A0A182FA84 A0A034VDF5 A0A1L8DMK3 T1DQ63 W5J2M0 A0A336M8B8 A0A182RFV3 A0A0K8WF15 A0A182XWJ0 A0A182PD55 A0A182QQB5 A0A182M5T7 A0A1Q3F6F7 A0A182VYD3 A0A182VP02 A0A182K5W5 A0A182WZI1 A0A182LQN9 Q7PTK7 A0A182HZM8 A0A182NMV3 B0X4C9 A0A182U039 E0VST7 A0A182IT76 Q16MM8 A0A310SHU0 A0A023ERT8 A0A1D2MN74 U5EVM9 A0A154PFD8 A0A164UAD1 A0A0P6GWU6 E9G055
Pubmed
19121390
26354079
23622113
22118469
20798317
28648823
+ More
20075255 24508170 30249741 21719571 18362917 19820115 21347285 23537049 28004739 17994087 22516182 27129103 15632085 24845553 25315136 17550304 25401762 26823975 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 20353571 26108605 25348373 20920257 23761445 25244985 20966253 12364791 14747013 17210077 20566863 17510324 24945155 26483478 27289101 21292972
20075255 24508170 30249741 21719571 18362917 19820115 21347285 23537049 28004739 17994087 22516182 27129103 15632085 24845553 25315136 17550304 25401762 26823975 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 20353571 26108605 25348373 20920257 23761445 25244985 20966253 12364791 14747013 17210077 20566863 17510324 24945155 26483478 27289101 21292972
EMBL
BABH01015062
KQ459603
KPI93123.1
ODYU01003698
SOQ42787.1
KQ460711
+ More
KPJ12686.1 GAIX01004274 JAA88286.1 AGBW02012899 OWR44246.1 GBYB01003718 JAG73485.1 GL437203 EFN70840.1 NNAY01000733 OXU26766.1 KK107231 QOIP01000009 EZA55066.1 RLU19016.1 KQ981042 KYN10077.1 GL887802 EGI70007.1 KQ976716 KYM76809.1 KQ971362 EFA07887.2 GL451577 EFN78978.1 ADTU01002228 KQ981253 KYN44172.1 KQ982431 KYQ56476.1 KQ414657 KOC65860.1 KZ288467 PBC25386.1 APGK01056846 KB741277 KB630186 KB632028 KB632155 ENN71166.1 ERL83459.1 ERL88126.1 ERL89221.1 GEZM01038750 JAV81529.1 CH940652 EDW59611.1 BT127639 AEE62601.1 KQ435698 KOX80757.1 KQ978344 KYM94825.1 CH964251 EDW83380.1 CH902617 EDV41713.1 CH933806 EDW16092.2 GECZ01031322 JAS38447.1 NEVH01004429 PNF39376.1 OUUW01000008 SPP83735.1 GDKW01001787 JAI54808.1 CM000070 EAL29195.1 KK852663 KDR19057.1 ACPB03015962 GAHY01000516 JAA76994.1 KA647916 AFP62545.1 GFTR01005477 JAW10949.1 CM000160 EDW96049.1 KK854009 PTY09218.1 CP012526 ALC46950.1 CH916373 EDV94919.1 GBHO01018273 GBHO01018272 GBRD01005465 GBRD01000941 GBRD01000939 GBRD01000938 GDHC01010120 JAG25331.1 JAG25332.1 JAG60356.1 JAQ08509.1 CH954181 EDV48470.1 CM000364 EDX12317.1 ABLF02023929 ABLF02023935 CH480840 EDW49451.1 AE014297 AY075171 AAF55688.1 AAL68041.1 GGMR01007747 MBY20366.1 GEBQ01006434 JAT33543.1 GAMC01014295 JAB92260.1 JXJN01018058 CCAG010009158 EZ423214 ADD19490.1 JRES01001410 KNC23140.1 GGFJ01006675 MBW55816.1 GGFM01001995 MBW22746.1 GGFK01003647 MBW36968.1 GAKP01018413 JAC40539.1 GFDF01006514 JAV07570.1 GAMD01002729 JAA98861.1 ADMH02002183 GGFL01004297 ETN57996.1 MBW68475.1 UFQT01000546 UFQT01001139 SSX25129.1 SSX29115.1 GDHF01002869 JAI49445.1 AXCN02001556 AXCM01004735 GFDL01011894 JAV23151.1 AAAB01008799 EAA03774.4 APCN01001987 DS232337 EDS40282.1 DS235756 EEB16443.1 AXCP01007799 CH477856 EAT35589.1 KQ762903 OAD55279.1 JXUM01054212 GAPW01002169 KQ561810 JAC11429.1 KXJ77457.1 LJIJ01000833 ODM94275.1 GANO01000951 JAB58920.1 KQ434893 KZC10573.1 LRGB01001581 KZS11186.1 GDIQ01027675 JAN67062.1 GL732528 EFX86841.1
KPJ12686.1 GAIX01004274 JAA88286.1 AGBW02012899 OWR44246.1 GBYB01003718 JAG73485.1 GL437203 EFN70840.1 NNAY01000733 OXU26766.1 KK107231 QOIP01000009 EZA55066.1 RLU19016.1 KQ981042 KYN10077.1 GL887802 EGI70007.1 KQ976716 KYM76809.1 KQ971362 EFA07887.2 GL451577 EFN78978.1 ADTU01002228 KQ981253 KYN44172.1 KQ982431 KYQ56476.1 KQ414657 KOC65860.1 KZ288467 PBC25386.1 APGK01056846 KB741277 KB630186 KB632028 KB632155 ENN71166.1 ERL83459.1 ERL88126.1 ERL89221.1 GEZM01038750 JAV81529.1 CH940652 EDW59611.1 BT127639 AEE62601.1 KQ435698 KOX80757.1 KQ978344 KYM94825.1 CH964251 EDW83380.1 CH902617 EDV41713.1 CH933806 EDW16092.2 GECZ01031322 JAS38447.1 NEVH01004429 PNF39376.1 OUUW01000008 SPP83735.1 GDKW01001787 JAI54808.1 CM000070 EAL29195.1 KK852663 KDR19057.1 ACPB03015962 GAHY01000516 JAA76994.1 KA647916 AFP62545.1 GFTR01005477 JAW10949.1 CM000160 EDW96049.1 KK854009 PTY09218.1 CP012526 ALC46950.1 CH916373 EDV94919.1 GBHO01018273 GBHO01018272 GBRD01005465 GBRD01000941 GBRD01000939 GBRD01000938 GDHC01010120 JAG25331.1 JAG25332.1 JAG60356.1 JAQ08509.1 CH954181 EDV48470.1 CM000364 EDX12317.1 ABLF02023929 ABLF02023935 CH480840 EDW49451.1 AE014297 AY075171 AAF55688.1 AAL68041.1 GGMR01007747 MBY20366.1 GEBQ01006434 JAT33543.1 GAMC01014295 JAB92260.1 JXJN01018058 CCAG010009158 EZ423214 ADD19490.1 JRES01001410 KNC23140.1 GGFJ01006675 MBW55816.1 GGFM01001995 MBW22746.1 GGFK01003647 MBW36968.1 GAKP01018413 JAC40539.1 GFDF01006514 JAV07570.1 GAMD01002729 JAA98861.1 ADMH02002183 GGFL01004297 ETN57996.1 MBW68475.1 UFQT01000546 UFQT01001139 SSX25129.1 SSX29115.1 GDHF01002869 JAI49445.1 AXCN02001556 AXCM01004735 GFDL01011894 JAV23151.1 AAAB01008799 EAA03774.4 APCN01001987 DS232337 EDS40282.1 DS235756 EEB16443.1 AXCP01007799 CH477856 EAT35589.1 KQ762903 OAD55279.1 JXUM01054212 GAPW01002169 KQ561810 JAC11429.1 KXJ77457.1 LJIJ01000833 ODM94275.1 GANO01000951 JAB58920.1 KQ434893 KZC10573.1 LRGB01001581 KZS11186.1 GDIQ01027675 JAN67062.1 GL732528 EFX86841.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000000311
UP000215335
+ More
UP000002358 UP000053097 UP000279307 UP000078492 UP000007755 UP000078540 UP000007266 UP000008237 UP000005205 UP000078541 UP000075809 UP000053825 UP000242457 UP000005203 UP000019118 UP000030742 UP000008792 UP000053105 UP000078542 UP000007798 UP000007801 UP000009192 UP000235965 UP000192223 UP000268350 UP000001819 UP000027135 UP000015103 UP000192221 UP000095301 UP000002282 UP000092553 UP000001070 UP000008711 UP000000304 UP000007819 UP000001292 UP000000803 UP000095300 UP000092443 UP000078200 UP000092460 UP000092445 UP000091820 UP000092444 UP000037069 UP000069272 UP000000673 UP000075900 UP000076408 UP000075885 UP000075886 UP000075883 UP000075920 UP000075903 UP000075881 UP000076407 UP000075882 UP000007062 UP000075840 UP000075884 UP000002320 UP000075902 UP000009046 UP000075880 UP000008820 UP000069940 UP000249989 UP000094527 UP000076502 UP000076858 UP000000305
UP000002358 UP000053097 UP000279307 UP000078492 UP000007755 UP000078540 UP000007266 UP000008237 UP000005205 UP000078541 UP000075809 UP000053825 UP000242457 UP000005203 UP000019118 UP000030742 UP000008792 UP000053105 UP000078542 UP000007798 UP000007801 UP000009192 UP000235965 UP000192223 UP000268350 UP000001819 UP000027135 UP000015103 UP000192221 UP000095301 UP000002282 UP000092553 UP000001070 UP000008711 UP000000304 UP000007819 UP000001292 UP000000803 UP000095300 UP000092443 UP000078200 UP000092460 UP000092445 UP000091820 UP000092444 UP000037069 UP000069272 UP000000673 UP000075900 UP000076408 UP000075885 UP000075886 UP000075883 UP000075920 UP000075903 UP000075881 UP000076407 UP000075882 UP000007062 UP000075840 UP000075884 UP000002320 UP000075902 UP000009046 UP000075880 UP000008820 UP000069940 UP000249989 UP000094527 UP000076502 UP000076858 UP000000305
Interpro
Gene 3D
ProteinModelPortal
H9JC86
A0A194PJA8
A0A2H1VPR7
A0A0N1IP67
S4P883
A0A212ERW3
+ More
A0A0C9PR51 E2A6X4 A0A232F7C7 K7IZ66 A0A026WHC5 A0A195DB47 F4W717 A0A195AXY8 D6WYF1 E2BZ72 A0A158NM95 A0A151K0R2 A0A151X7X2 A0A0L7R4R3 A0A2A3E297 A0A088AQW7 N6TS78 A0A1Y1M6R3 B4M4Q0 J3JWM1 A0A0M9AAV2 A0A195C2K7 B4NF05 B3LXQ6 B4K4T9 A0A1B6EKJ2 A0A2J7REX1 A0A1W4WRG1 A0A3B0JNL9 A0A0N7Z918 Q293L9 A0A067R6W0 R4G4S6 A0A1W4W8E4 T1PGL0 A0A224XSJ5 B4PNI6 A0A2R7VPY5 A0A0M4F532 B4JT29 A0A0A9Y113 B3P096 B4QSV7 J9JW82 B4IHT4 Q9VDV2 A0A2S2NUG2 A0A1B6MC88 A0A1I8PZ11 W8ATU1 A0A1A9YSC9 A0A1A9VTC3 A0A1B0BPK8 A0A1A9ZPP7 A0A1A9WTX5 D3TP62 A0A0L0BST2 A0A2M4BRW0 A0A2M3Z2K6 A0A2M4A8M5 A0A182FA84 A0A034VDF5 A0A1L8DMK3 T1DQ63 W5J2M0 A0A336M8B8 A0A182RFV3 A0A0K8WF15 A0A182XWJ0 A0A182PD55 A0A182QQB5 A0A182M5T7 A0A1Q3F6F7 A0A182VYD3 A0A182VP02 A0A182K5W5 A0A182WZI1 A0A182LQN9 Q7PTK7 A0A182HZM8 A0A182NMV3 B0X4C9 A0A182U039 E0VST7 A0A182IT76 Q16MM8 A0A310SHU0 A0A023ERT8 A0A1D2MN74 U5EVM9 A0A154PFD8 A0A164UAD1 A0A0P6GWU6 E9G055
A0A0C9PR51 E2A6X4 A0A232F7C7 K7IZ66 A0A026WHC5 A0A195DB47 F4W717 A0A195AXY8 D6WYF1 E2BZ72 A0A158NM95 A0A151K0R2 A0A151X7X2 A0A0L7R4R3 A0A2A3E297 A0A088AQW7 N6TS78 A0A1Y1M6R3 B4M4Q0 J3JWM1 A0A0M9AAV2 A0A195C2K7 B4NF05 B3LXQ6 B4K4T9 A0A1B6EKJ2 A0A2J7REX1 A0A1W4WRG1 A0A3B0JNL9 A0A0N7Z918 Q293L9 A0A067R6W0 R4G4S6 A0A1W4W8E4 T1PGL0 A0A224XSJ5 B4PNI6 A0A2R7VPY5 A0A0M4F532 B4JT29 A0A0A9Y113 B3P096 B4QSV7 J9JW82 B4IHT4 Q9VDV2 A0A2S2NUG2 A0A1B6MC88 A0A1I8PZ11 W8ATU1 A0A1A9YSC9 A0A1A9VTC3 A0A1B0BPK8 A0A1A9ZPP7 A0A1A9WTX5 D3TP62 A0A0L0BST2 A0A2M4BRW0 A0A2M3Z2K6 A0A2M4A8M5 A0A182FA84 A0A034VDF5 A0A1L8DMK3 T1DQ63 W5J2M0 A0A336M8B8 A0A182RFV3 A0A0K8WF15 A0A182XWJ0 A0A182PD55 A0A182QQB5 A0A182M5T7 A0A1Q3F6F7 A0A182VYD3 A0A182VP02 A0A182K5W5 A0A182WZI1 A0A182LQN9 Q7PTK7 A0A182HZM8 A0A182NMV3 B0X4C9 A0A182U039 E0VST7 A0A182IT76 Q16MM8 A0A310SHU0 A0A023ERT8 A0A1D2MN74 U5EVM9 A0A154PFD8 A0A164UAD1 A0A0P6GWU6 E9G055
PDB
4A9A
E-value=1.22249e-96,
Score=901
Ontologies
KEGG
GO
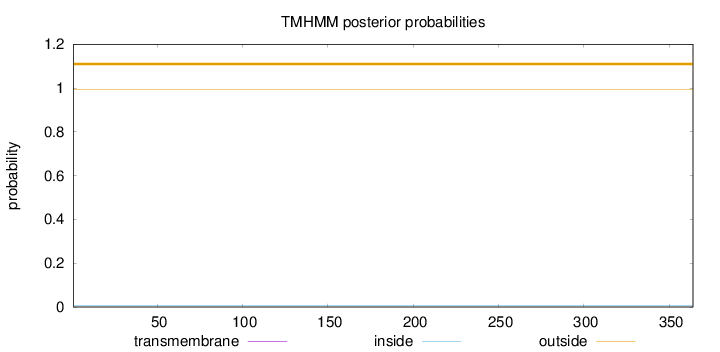
Topology
Length:
364
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00392000000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00554
outside
1 - 364
Population Genetic Test Statistics
Pi
30.801663
Theta
25.41199
Tajima's D
0.521489
CLR
0.922052
CSRT
0.525723713814309
Interpretation
Uncertain
