Pre Gene Modal
BGIBMGA006984
Annotation
G10_protein_[Bombyx_mori]
Full name
Protein BUD31 homolog
Alternative Name
Protein G10 homolog
Location in the cell
Nuclear Reliability : 3.077
Sequence
CDS
ATGCCTAAGGTTCGTCGCAGTAGGAAACCACCACCAGAAGGATGGGAATTAATTGAGCCAACACTTGAGGAACTTGAACAGAAAATGCGAGAAGCCGAAACTGAACCGCACGAAGGGAAACGAAAACAAGAATCCTTATGGCCAATTTTCAAAATACATCATCAGAAATCGCGTTACATCTATGACTTATTTTACAGACGTAAGGCCATTAGTAGAGAACTTTACCAATATTGCTTAAATGAAAAAATAGCTGACGCCAATCTTATAGCGAAATGGAAAAAGACTGGGTATGAAAATTTGTGTTGTCTTCGCTGCATACAAACTAGAGATACAAATTTTGCAACTAATTGTATATGTAGAGTTCCGAAGAGTAAGCTGGAAGAGGGAAGGATTGTTGAATGTGTCCATTGTGGTTGCAGAGGTTGTTCAGGATAG
Protein
MPKVRRSRKPPPEGWELIEPTLEELEQKMREAETEPHEGKRKQESLWPIFKIHHQKSRYIYDLFYRRKAISRELYQYCLNEKIADANLIAKWKKTGYENLCCLRCIQTRDTNFATNCICRVPKSKLEEGRIVECVHCGCRGCSG
Summary
Similarity
Belongs to the BUD31 (G10) family.
Keywords
Nucleus
Feature
chain Protein BUD31 homolog
Uniprot
Q2F698
S4NWW0
A0A212F860
A0A2A4K629
A0A2H1VBF8
A0A0L7LL69
+ More
A0A0N1IDL1 A0A194PPQ6 W8BVT2 A0A0K8WK25 A0A034WKN3 A0A0L0CML9 A0A0A1WMU2 A0A1I8N8S2 A0A1I8P1X7 B4M2P1 A0A195C794 A0A195DJP3 F4WK43 A0A151I4N6 A0A0J7KB83 E2BKK6 A0A158NU22 A0A151WZF8 A0A026X0P9 A0A0M3QZF8 A0A1B0FGZ7 A0A1A9XIT4 A0A1A9UUK4 A0A1B0AX87 A0A1A9ZBH4 A0A0L7RD38 A0A154P1G5 A0A310SPP5 B4KUF5 A0A336LD97 A0A1A9WPL9 A0A182UNE4 A0A1W4UN43 B4LIW4 B4PYD1 A0A0P9AB08 B3NV82 U5EVQ0 B4L4G4 B4JJM4 A0A182W2M1 E1ZVB2 A0A0K8TQZ2 A0A182Q1U6 A0A182XVE4 A0A182MT94 A0A182K797 A0A182LFN2 A0A182XLZ2 Q7QBT5 A0A182R755 A0A182HTH5 B0WMX4 Q170V3 A0A023EGF6 A0A2M3ZCP0 A0A182NG91 A0A2M4ATN6 A0A2M4C2S2 W5JES2 A0A182FQ02 B4N2J1 A0A1Q3F535 A0A182PEM1 A0A1B0DKV3 A0A1B0CN81 B4R2S6 B4IE04 O97454 A0A0M9ABR5 A0A1L8DH03 C3Y9A1 Q962X9 E0VGI9 B4JVS3 A0A3B0JEC2 B5DYK6 B4GNU7 J3JY95 A0A195FJ20 A0A1Y1KII8 A0A1B6KSX0 A0A1B6FZM9 A0A1B6ILB9 A0A0A9YNU3 A0A0A9YT74 Q962X2 A0A067QSW3 A0A1B6DI33 A0A0P4WS20 C1BSC6 A0A3R7QYM9 A0A2A3E274 C1C0M3
A0A0N1IDL1 A0A194PPQ6 W8BVT2 A0A0K8WK25 A0A034WKN3 A0A0L0CML9 A0A0A1WMU2 A0A1I8N8S2 A0A1I8P1X7 B4M2P1 A0A195C794 A0A195DJP3 F4WK43 A0A151I4N6 A0A0J7KB83 E2BKK6 A0A158NU22 A0A151WZF8 A0A026X0P9 A0A0M3QZF8 A0A1B0FGZ7 A0A1A9XIT4 A0A1A9UUK4 A0A1B0AX87 A0A1A9ZBH4 A0A0L7RD38 A0A154P1G5 A0A310SPP5 B4KUF5 A0A336LD97 A0A1A9WPL9 A0A182UNE4 A0A1W4UN43 B4LIW4 B4PYD1 A0A0P9AB08 B3NV82 U5EVQ0 B4L4G4 B4JJM4 A0A182W2M1 E1ZVB2 A0A0K8TQZ2 A0A182Q1U6 A0A182XVE4 A0A182MT94 A0A182K797 A0A182LFN2 A0A182XLZ2 Q7QBT5 A0A182R755 A0A182HTH5 B0WMX4 Q170V3 A0A023EGF6 A0A2M3ZCP0 A0A182NG91 A0A2M4ATN6 A0A2M4C2S2 W5JES2 A0A182FQ02 B4N2J1 A0A1Q3F535 A0A182PEM1 A0A1B0DKV3 A0A1B0CN81 B4R2S6 B4IE04 O97454 A0A0M9ABR5 A0A1L8DH03 C3Y9A1 Q962X9 E0VGI9 B4JVS3 A0A3B0JEC2 B5DYK6 B4GNU7 J3JY95 A0A195FJ20 A0A1Y1KII8 A0A1B6KSX0 A0A1B6FZM9 A0A1B6ILB9 A0A0A9YNU3 A0A0A9YT74 Q962X2 A0A067QSW3 A0A1B6DI33 A0A0P4WS20 C1BSC6 A0A3R7QYM9 A0A2A3E274 C1C0M3
Pubmed
19121390
23622113
22118469
26227816
26354079
24495485
+ More
25348373 26108605 25830018 25315136 17994087 21719571 20798317 21347285 24508170 30249741 18057021 17550304 26369729 25244985 20966253 12364791 14747013 17210077 17510324 24945155 26483478 20920257 23761445 9671597 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18563158 20566863 15632085 22516182 28004739 25401762 24845553
25348373 26108605 25830018 25315136 17994087 21719571 20798317 21347285 24508170 30249741 18057021 17550304 26369729 25244985 20966253 12364791 14747013 17210077 17510324 24945155 26483478 20920257 23761445 9671597 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18563158 20566863 15632085 22516182 28004739 25401762 24845553
EMBL
BABH01015049
DQ311174
ABD36119.1
GAIX01012397
JAA80163.1
AGBW02009792
+ More
AGBW02008359 OWR49913.1 OWR53590.1 NWSH01000131 PCG79223.1 ODYU01001480 SOQ37732.1 JTDY01000700 JTDY01000361 KOB76172.1 KOB77545.1 KQ460711 KPJ12678.1 KQ459603 KPI93115.1 GAMC01003223 JAC03333.1 GDHF01000801 JAI51513.1 GAKP01002811 JAC56141.1 JRES01000290 KNC32689.1 GBXI01014549 JAC99742.1 CH940651 EDW65945.1 KQ978143 KYM96744.1 KQ980794 KYN13071.1 GL888193 EGI65415.1 KQ976445 KYM86040.1 LBMM01010179 KMQ87628.1 GL448810 EFN83793.1 ADTU01026246 KQ982638 KYQ53284.1 KK107054 QOIP01000004 EZA61628.1 RLU23963.1 CP012528 ALC49299.1 CCAG010009006 JXJN01005127 KQ414615 KOC68773.1 KQ434796 KZC05701.1 KQ760382 OAD60676.1 CH933808 EDW10742.1 KRG05454.1 UFQS01001708 UFQS01002288 UFQT01001708 UFQT01002288 SSX11914.1 SSX13688.1 SSX31476.1 CH940648 EDW60414.1 CM000162 EDX01993.1 CH902625 KPU75449.1 CH954180 EDV46070.1 GANO01000920 JAB58951.1 CH933810 EDW07442.1 CH916370 EDV99776.1 GL434491 EFN74888.1 GDAI01000821 JAI16782.1 AXCN02001628 AXCM01000235 AAAB01008859 EAA07460.1 APCN01000528 DS232003 EDS31366.1 CH477466 EAT40484.1 JXUM01045219 GAPW01005678 KQ561430 JAC07920.1 KXJ78609.1 GGFM01005520 MBW26271.1 GGFK01010813 MBW44134.1 GGFJ01010408 MBW59549.1 ADMH02001428 ETN62571.1 CH963925 EDW78580.1 GFDL01012379 JAV22666.1 AJVK01016125 AJWK01019879 CM000366 EDX17625.1 CH480830 EDW45812.1 AF046044 AF045587 AE014298 AY118623 KX532010 AAC97603.1 AAC98483.1 AAF48023.1 AAM49992.1 ANY27820.1 KQ435889 KQ435689 KOX69494.1 KOX81295.1 GFDF01008383 JAV05701.1 GG666492 EEN63157.1 AF395865 DS235145 EEB12495.1 CH916375 EDV98061.1 OUUW01000005 SPP80734.1 CM000070 EDY67623.1 CH479186 EDW38830.1 BT128219 AEE63180.1 KQ981522 KYN40660.1 GEZM01087592 GEZM01087591 JAV58597.1 GEBQ01025459 JAT14518.1 GECZ01014127 JAS55642.1 GECU01019993 GECU01005031 JAS87713.1 JAT02676.1 GBHO01010831 JAG32773.1 GBHO01010834 GBRD01005453 JAG32770.1 JAG60368.1 AF397147 AAK84395.1 KK852980 KDR12968.1 GEDC01031390 GEDC01011952 JAS05908.1 JAS25346.1 GDRN01036637 JAI67333.1 BT077505 HACA01012197 ACO11929.1 CDW29558.1 QCYY01000661 ROT83611.1 KZ288433 PBC25795.1 BT080402 ACO14826.1
AGBW02008359 OWR49913.1 OWR53590.1 NWSH01000131 PCG79223.1 ODYU01001480 SOQ37732.1 JTDY01000700 JTDY01000361 KOB76172.1 KOB77545.1 KQ460711 KPJ12678.1 KQ459603 KPI93115.1 GAMC01003223 JAC03333.1 GDHF01000801 JAI51513.1 GAKP01002811 JAC56141.1 JRES01000290 KNC32689.1 GBXI01014549 JAC99742.1 CH940651 EDW65945.1 KQ978143 KYM96744.1 KQ980794 KYN13071.1 GL888193 EGI65415.1 KQ976445 KYM86040.1 LBMM01010179 KMQ87628.1 GL448810 EFN83793.1 ADTU01026246 KQ982638 KYQ53284.1 KK107054 QOIP01000004 EZA61628.1 RLU23963.1 CP012528 ALC49299.1 CCAG010009006 JXJN01005127 KQ414615 KOC68773.1 KQ434796 KZC05701.1 KQ760382 OAD60676.1 CH933808 EDW10742.1 KRG05454.1 UFQS01001708 UFQS01002288 UFQT01001708 UFQT01002288 SSX11914.1 SSX13688.1 SSX31476.1 CH940648 EDW60414.1 CM000162 EDX01993.1 CH902625 KPU75449.1 CH954180 EDV46070.1 GANO01000920 JAB58951.1 CH933810 EDW07442.1 CH916370 EDV99776.1 GL434491 EFN74888.1 GDAI01000821 JAI16782.1 AXCN02001628 AXCM01000235 AAAB01008859 EAA07460.1 APCN01000528 DS232003 EDS31366.1 CH477466 EAT40484.1 JXUM01045219 GAPW01005678 KQ561430 JAC07920.1 KXJ78609.1 GGFM01005520 MBW26271.1 GGFK01010813 MBW44134.1 GGFJ01010408 MBW59549.1 ADMH02001428 ETN62571.1 CH963925 EDW78580.1 GFDL01012379 JAV22666.1 AJVK01016125 AJWK01019879 CM000366 EDX17625.1 CH480830 EDW45812.1 AF046044 AF045587 AE014298 AY118623 KX532010 AAC97603.1 AAC98483.1 AAF48023.1 AAM49992.1 ANY27820.1 KQ435889 KQ435689 KOX69494.1 KOX81295.1 GFDF01008383 JAV05701.1 GG666492 EEN63157.1 AF395865 DS235145 EEB12495.1 CH916375 EDV98061.1 OUUW01000005 SPP80734.1 CM000070 EDY67623.1 CH479186 EDW38830.1 BT128219 AEE63180.1 KQ981522 KYN40660.1 GEZM01087592 GEZM01087591 JAV58597.1 GEBQ01025459 JAT14518.1 GECZ01014127 JAS55642.1 GECU01019993 GECU01005031 JAS87713.1 JAT02676.1 GBHO01010831 JAG32773.1 GBHO01010834 GBRD01005453 JAG32770.1 JAG60368.1 AF397147 AAK84395.1 KK852980 KDR12968.1 GEDC01031390 GEDC01011952 JAS05908.1 JAS25346.1 GDRN01036637 JAI67333.1 BT077505 HACA01012197 ACO11929.1 CDW29558.1 QCYY01000661 ROT83611.1 KZ288433 PBC25795.1 BT080402 ACO14826.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000037510
UP000053240
UP000053268
+ More
UP000037069 UP000095301 UP000095300 UP000008792 UP000078542 UP000078492 UP000007755 UP000078540 UP000036403 UP000008237 UP000005205 UP000075809 UP000053097 UP000279307 UP000092553 UP000092444 UP000092443 UP000078200 UP000092460 UP000092445 UP000053825 UP000076502 UP000009192 UP000091820 UP000075903 UP000192221 UP000002282 UP000007801 UP000008711 UP000001070 UP000075920 UP000000311 UP000075886 UP000076408 UP000075883 UP000075881 UP000075882 UP000076407 UP000007062 UP000075900 UP000075840 UP000002320 UP000008820 UP000069940 UP000249989 UP000075884 UP000000673 UP000069272 UP000007798 UP000075885 UP000092462 UP000092461 UP000000304 UP000001292 UP000000803 UP000053105 UP000001554 UP000009046 UP000268350 UP000001819 UP000008744 UP000078541 UP000027135 UP000283509 UP000242457
UP000037069 UP000095301 UP000095300 UP000008792 UP000078542 UP000078492 UP000007755 UP000078540 UP000036403 UP000008237 UP000005205 UP000075809 UP000053097 UP000279307 UP000092553 UP000092444 UP000092443 UP000078200 UP000092460 UP000092445 UP000053825 UP000076502 UP000009192 UP000091820 UP000075903 UP000192221 UP000002282 UP000007801 UP000008711 UP000001070 UP000075920 UP000000311 UP000075886 UP000076408 UP000075883 UP000075881 UP000075882 UP000076407 UP000007062 UP000075900 UP000075840 UP000002320 UP000008820 UP000069940 UP000249989 UP000075884 UP000000673 UP000069272 UP000007798 UP000075885 UP000092462 UP000092461 UP000000304 UP000001292 UP000000803 UP000053105 UP000001554 UP000009046 UP000268350 UP000001819 UP000008744 UP000078541 UP000027135 UP000283509 UP000242457
Pfam
PF01125 G10
ProteinModelPortal
Q2F698
S4NWW0
A0A212F860
A0A2A4K629
A0A2H1VBF8
A0A0L7LL69
+ More
A0A0N1IDL1 A0A194PPQ6 W8BVT2 A0A0K8WK25 A0A034WKN3 A0A0L0CML9 A0A0A1WMU2 A0A1I8N8S2 A0A1I8P1X7 B4M2P1 A0A195C794 A0A195DJP3 F4WK43 A0A151I4N6 A0A0J7KB83 E2BKK6 A0A158NU22 A0A151WZF8 A0A026X0P9 A0A0M3QZF8 A0A1B0FGZ7 A0A1A9XIT4 A0A1A9UUK4 A0A1B0AX87 A0A1A9ZBH4 A0A0L7RD38 A0A154P1G5 A0A310SPP5 B4KUF5 A0A336LD97 A0A1A9WPL9 A0A182UNE4 A0A1W4UN43 B4LIW4 B4PYD1 A0A0P9AB08 B3NV82 U5EVQ0 B4L4G4 B4JJM4 A0A182W2M1 E1ZVB2 A0A0K8TQZ2 A0A182Q1U6 A0A182XVE4 A0A182MT94 A0A182K797 A0A182LFN2 A0A182XLZ2 Q7QBT5 A0A182R755 A0A182HTH5 B0WMX4 Q170V3 A0A023EGF6 A0A2M3ZCP0 A0A182NG91 A0A2M4ATN6 A0A2M4C2S2 W5JES2 A0A182FQ02 B4N2J1 A0A1Q3F535 A0A182PEM1 A0A1B0DKV3 A0A1B0CN81 B4R2S6 B4IE04 O97454 A0A0M9ABR5 A0A1L8DH03 C3Y9A1 Q962X9 E0VGI9 B4JVS3 A0A3B0JEC2 B5DYK6 B4GNU7 J3JY95 A0A195FJ20 A0A1Y1KII8 A0A1B6KSX0 A0A1B6FZM9 A0A1B6ILB9 A0A0A9YNU3 A0A0A9YT74 Q962X2 A0A067QSW3 A0A1B6DI33 A0A0P4WS20 C1BSC6 A0A3R7QYM9 A0A2A3E274 C1C0M3
A0A0N1IDL1 A0A194PPQ6 W8BVT2 A0A0K8WK25 A0A034WKN3 A0A0L0CML9 A0A0A1WMU2 A0A1I8N8S2 A0A1I8P1X7 B4M2P1 A0A195C794 A0A195DJP3 F4WK43 A0A151I4N6 A0A0J7KB83 E2BKK6 A0A158NU22 A0A151WZF8 A0A026X0P9 A0A0M3QZF8 A0A1B0FGZ7 A0A1A9XIT4 A0A1A9UUK4 A0A1B0AX87 A0A1A9ZBH4 A0A0L7RD38 A0A154P1G5 A0A310SPP5 B4KUF5 A0A336LD97 A0A1A9WPL9 A0A182UNE4 A0A1W4UN43 B4LIW4 B4PYD1 A0A0P9AB08 B3NV82 U5EVQ0 B4L4G4 B4JJM4 A0A182W2M1 E1ZVB2 A0A0K8TQZ2 A0A182Q1U6 A0A182XVE4 A0A182MT94 A0A182K797 A0A182LFN2 A0A182XLZ2 Q7QBT5 A0A182R755 A0A182HTH5 B0WMX4 Q170V3 A0A023EGF6 A0A2M3ZCP0 A0A182NG91 A0A2M4ATN6 A0A2M4C2S2 W5JES2 A0A182FQ02 B4N2J1 A0A1Q3F535 A0A182PEM1 A0A1B0DKV3 A0A1B0CN81 B4R2S6 B4IE04 O97454 A0A0M9ABR5 A0A1L8DH03 C3Y9A1 Q962X9 E0VGI9 B4JVS3 A0A3B0JEC2 B5DYK6 B4GNU7 J3JY95 A0A195FJ20 A0A1Y1KII8 A0A1B6KSX0 A0A1B6FZM9 A0A1B6ILB9 A0A0A9YNU3 A0A0A9YT74 Q962X2 A0A067QSW3 A0A1B6DI33 A0A0P4WS20 C1BSC6 A0A3R7QYM9 A0A2A3E274 C1C0M3
PDB
6QDV
E-value=8.83405e-66,
Score=629
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
Length:
144
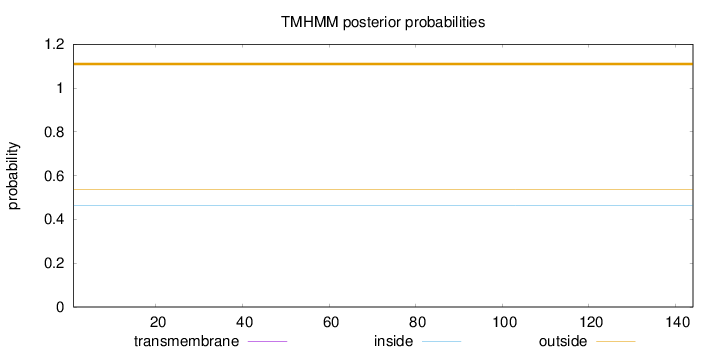
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.46222
outside
1 - 144
Population Genetic Test Statistics
Pi
230.607294
Theta
207.904172
Tajima's D
0.339313
CLR
144.89827
CSRT
0.463576821158942
Interpretation
Uncertain
