Gene
KWMTBOMO10516
Pre Gene Modal
BGIBMGA007125
Annotation
innexin_4_[Bombyx_mori]
Full name
Innexin
+ More
Innexin inx2
Innexin inx2
Alternative Name
Gap junction protein prp33
Pas-related protein 33
G-Inx2
Pas-related protein 33
G-Inx2
Location in the cell
PlasmaMembrane Reliability : 4.335
Sequence
CDS
ATGATTGATCTTTTTATGCCTTTTCGTTCATTTTTGAAATTTGAGAATGTATGTACTGATAATAATATTTTTCGAATGCATTACAAGCTCACAGTCATAATATTATTGGTGTTTACACTTCTTGTGACTTCAAAGCAGTTTTTCGGAGAGCCCATACATTGTATGAGCGGCAATGATAAAGGCAATGATAAGGACGCCGTGAACAGTTATTGTTGGATATATGGAACATACACGCTGAAGAGCCAGCTTTTAGGTGTTGAAGGCAGGCACATGGCATATGTAGGAGTAGGACCGGCTAAGTCCGACGATGATGAGCAGATCAAACACACTTACTACCAGTGGGTGTGTTTTGTACTACTTGGTCAAGCCACCATGTTTTATGCTCCTCGATACCTGTGGAAAATGTGGGAAGGAGGCAGGCTGAAGGCACTGGCAGCTGATCTTAGCAGTCCAATGGTTTCGAAGGATTGGTCAGAGTTCAGGCGCAAGGAACTTGTCTCATATTTTAATTACACAAATATGTATACTCATAATATGTATGCCCTGCGTTATGCATTTTGTGAGTTACTGAACCTTGTAAATGTGGTTGGTCAAATATTTATTCTGGACTTGTTCCTTGGTGGTTCATTCCGTAACTATGGTGCTGCAGTTGCAGCTTTTACGCACACCCCACGCATACCTAACGATTTTACCAACTTCACCTCTGTCAACCCCATGGATGAATTTTTTCCCAAACTCACTAAATGCTGGTTAAGAAACTATGGACCTTCAGGCTCTCTTGAATTAAAGGATCGATTGTGTGTACTACCTTTAAATATTGTTAATGAAAAGATATTTGTTATCCTTTGGTTCTGGCTTATTATATTAACGGCTTTTTCGATACTAGCAGTAGTTTTTCGTTTTCTCTTGCTTATATTATATCCACTGCGGACAGTAATGATACGTGGACAAATACGCTATGTTAAAAGAAGTGTTGTCTCACGTATTGTAAAGAGATTTGGTTTTGGGGACTGGTTTATTCTCCACTTATTGGGTAAGAACATGAATCCAATAATTTTTAAGGATTTAGTACTTGAATTGGCCAAAGAAATAGAACATAATGCTATAATGGTATAA
Protein
MIDLFMPFRSFLKFENVCTDNNIFRMHYKLTVIILLVFTLLVTSKQFFGEPIHCMSGNDKGNDKDAVNSYCWIYGTYTLKSQLLGVEGRHMAYVGVGPAKSDDDEQIKHTYYQWVCFVLLGQATMFYAPRYLWKMWEGGRLKALAADLSSPMVSKDWSEFRRKELVSYFNYTNMYTHNMYALRYAFCELLNLVNVVGQIFILDLFLGGSFRNYGAAVAAFTHTPRIPNDFTNFTSVNPMDEFFPKLTKCWLRNYGPSGSLELKDRLCVLPLNIVNEKIFVILWFWLIILTAFSILAVVFRFLLLILYPLRTVMIRGQIRYVKRSVVSRIVKRFGFGDWFILHLLGKNMNPIIFKDLVLELAKEIEHNAIMV
Summary
Description
Structural component of the gap junctions.
Structural components of the gap junctions. Involved in gap junctional communication between germline and somatic cells which is essential for normal oogenesis. In embryonic epidermis, required for epithelial morphogenesis. Required for keyhole formation during early stages of proventriculus development in response to wg signaling. In follicle cells, promotes the formation of egg chambers in part through regulation of shg and baz at the boundary between germ cells and follicle cells. In inner germarial sheath cells, required for survival of early germ cells and for cyst formation.
Structural components of the gap junctions. Involved in gap junctional communication between germline and somatic cells which is essential for normal oogenesis. In embryonic epidermis, required for epithelial morphogenesis. Required for keyhole formation during early stages of proventriculus development in response to wg signaling. In follicle cells, promotes the formation of egg chambers in part through regulation of shg and baz at the boundary between germ cells and follicle cells. In inner germarial sheath cells, required for survival of early germ cells and for cyst formation.
Subunit
Monomer and heterooligomer with ogre or Inx3 (via cytoplasmic C-terminal region). Interacts (via cytoplasmic loop) with shg (via cytoplasmic region). Interacts with arm.
Similarity
Belongs to the pannexin family.
Keywords
Cell junction
Cell membrane
Complete proteome
Cytoplasm
Developmental protein
Differentiation
Gap junction
Ion channel
Ion transport
Membrane
Oogenesis
Reference proteome
Transmembrane
Transmembrane helix
Transport
Feature
chain Innexin
Uniprot
Q5XLD8
A2TEZ1
A0A2H1VAA9
A0A2A4K6C4
S4P161
H9JC80
+ More
A0A2A4K4P2 A0A0N1PHR1 A0A194PK59 E2BKJ4 A0A151WZW3 A0A151I4W9 F4WK32 A0A195DJR1 A0A3L8DU44 A0A195FL04 E1ZVA0 A0A026X0B1 A0A034VKH4 A0A0A9VUC1 A0A0K8VKP7 A0A1B6CKW9 A0A2J7PHC1 A0A182JZM3 A0A182VWE0 A0A195C9G7 A0A1A9XRV8 A0A1B0AP71 A2TEZ0 Q6SA04 A0A1B0G4W2 A0A1A9UUN2 A0A1A9ZE59 A0A182YB92 A0A182NA24 A0A194PPJ2 A0A0N0PCR1 W8B342 W8AZ10 V5GVY7 A0A1B0DD12 A0A182M026 A0A2P8YZB7 A0A0A1XMB6 E1JJF3 Q9V427 Q6XZI1 B3NXY3 Q6PYX4 A0A182R506 A0A1L8E3F2 A0A067R1S3 T1PEY5 A0A1B0CVA1 B4Q160 B4M9W2 Q9XYN1 A0A1I8P411 C0KLS6 A0A182QM11 A0A0L0BUG1 K9RY87 K9S0V4 A0A2A4K0K9 A0A2R7WYH0 A0A1A9WPK2 A0A1B6L2T6 B4IHI6 A0A084W2Z8 A0A158NU57 B3MQ11 U5EUV3 A0A3S2LJH0 B4NEG0 A0A3B0K691 A0A0P6IZ10 A0A182SMM5 A0A226EJI1 B4JMT1 A0A1B6IFW0 A0A212EYK5 A0A182GB47 A0A1W4WZU8 A0A1W4V8D0 D6X4J0 T1E2R6 A0A1E1WFT8 A0A1B6GUR0 Q29GY8 A0A224XSQ0 A0A069DSD1 Q16FA2 S4PA74 T1DI93 A0A0M5J2R8 A0A1J1I1G9 J3JY90 E1ZXE9 B4L565
A0A2A4K4P2 A0A0N1PHR1 A0A194PK59 E2BKJ4 A0A151WZW3 A0A151I4W9 F4WK32 A0A195DJR1 A0A3L8DU44 A0A195FL04 E1ZVA0 A0A026X0B1 A0A034VKH4 A0A0A9VUC1 A0A0K8VKP7 A0A1B6CKW9 A0A2J7PHC1 A0A182JZM3 A0A182VWE0 A0A195C9G7 A0A1A9XRV8 A0A1B0AP71 A2TEZ0 Q6SA04 A0A1B0G4W2 A0A1A9UUN2 A0A1A9ZE59 A0A182YB92 A0A182NA24 A0A194PPJ2 A0A0N0PCR1 W8B342 W8AZ10 V5GVY7 A0A1B0DD12 A0A182M026 A0A2P8YZB7 A0A0A1XMB6 E1JJF3 Q9V427 Q6XZI1 B3NXY3 Q6PYX4 A0A182R506 A0A1L8E3F2 A0A067R1S3 T1PEY5 A0A1B0CVA1 B4Q160 B4M9W2 Q9XYN1 A0A1I8P411 C0KLS6 A0A182QM11 A0A0L0BUG1 K9RY87 K9S0V4 A0A2A4K0K9 A0A2R7WYH0 A0A1A9WPK2 A0A1B6L2T6 B4IHI6 A0A084W2Z8 A0A158NU57 B3MQ11 U5EUV3 A0A3S2LJH0 B4NEG0 A0A3B0K691 A0A0P6IZ10 A0A182SMM5 A0A226EJI1 B4JMT1 A0A1B6IFW0 A0A212EYK5 A0A182GB47 A0A1W4WZU8 A0A1W4V8D0 D6X4J0 T1E2R6 A0A1E1WFT8 A0A1B6GUR0 Q29GY8 A0A224XSQ0 A0A069DSD1 Q16FA2 S4PA74 T1DI93 A0A0M5J2R8 A0A1J1I1G9 J3JY90 E1ZXE9 B4L565
Pubmed
17950307
23622113
19121390
26354079
20798317
21719571
+ More
30249741 24508170 25348373 25401762 26823975 25244985 24495485 29403074 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 10352230 10888681 12537569 11956317 11960713 15047872 16436513 19038051 22001874 17994087 18057021 28433752 24845553 25315136 17550304 10079517 26108605 24438588 21347285 22118469 26483478 18362917 19820115 24330624 15632085 26334808 17510324 25585357 22516182
30249741 24508170 25348373 25401762 26823975 25244985 24495485 29403074 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 10352230 10888681 12537569 11956317 11960713 15047872 16436513 19038051 22001874 17994087 18057021 28433752 24845553 25315136 17550304 10079517 26108605 24438588 21347285 22118469 26483478 18362917 19820115 24330624 15632085 26334808 17510324 25585357 22516182
EMBL
AY754727
AAV28707.1
EF197892
ABM88791.1
ODYU01001480
SOQ37731.1
+ More
NWSH01000131 PCG79222.1 GAIX01009046 JAA83514.1 BABH01015047 BABH01015048 BABH01015049 PCG79221.1 KQ460711 KPJ12677.1 KQ459603 KPI93114.1 GL448810 EFN83781.1 KQ982638 KYQ53297.1 KQ976445 KYM86057.1 GL888193 EGI65404.1 KQ980794 KYN13057.1 QOIP01000004 RLU23891.1 KQ981522 KYN40674.1 GL434491 EFN74876.1 KK107046 EZA61697.1 GAKP01016340 JAC42612.1 GBHO01044300 GBHO01030105 GBRD01012033 GDHC01002278 JAF99303.1 JAG13499.1 JAG53791.1 JAQ16351.1 GDHF01015923 GDHF01012857 JAI36391.1 JAI39457.1 GEDC01023218 GEDC01018802 JAS14080.1 JAS18496.1 NEVH01025142 PNF15718.1 KQ978143 KYM96758.1 JXJN01001234 EF197891 ABM88790.1 BABH01003247 AY461704 AAR97567.2 CCAG010000780 KQ459596 KPI95361.1 KQ460497 KPJ14319.1 GAMC01015097 JAB91458.1 GAMC01015098 JAB91457.1 GALX01004058 JAB64408.1 AJVK01014295 AXCM01003927 PYGN01000272 PSN49595.1 GBXI01002190 JAD12102.1 AE014298 ACZ95220.1 AF137269 AF172257 AY060368 AY196138 ODYU01003264 AAP40732.1 SOQ41789.1 CH954180 EDV47434.1 AY570724 AAS77384.1 GFDF01000920 JAV13164.1 KK853092 KDR11527.1 KA646715 AFP61344.1 AJWK01030336 CM000162 EDX02415.1 CH940655 EDW66021.1 AF115854 FJ619503 ACN41954.1 AXCN02001524 JRES01001325 KNC23628.1 KC018471 AFY62975.1 KC018473 AFY62977.1 NWSH01000281 PCG77801.1 KK855997 PTY24583.1 GEBQ01022017 JAT17960.1 CH480839 EDW49337.1 ATLV01019742 KE525279 KFB44592.1 ADTU01026241 CH902621 EDV44437.1 GANO01001288 JAB58583.1 RSAL01000001 RVE55224.1 CH964239 EDW82129.1 OUUW01000016 SPP88803.1 GDIQ01002289 JAN92448.1 LNIX01000003 OXA57277.1 CH916371 EDV92024.1 GECU01021896 JAS85810.1 AGBW02011483 OWR46576.1 JXUM01157673 KQ572819 KXJ67988.1 KQ971380 EEZ97563.1 GALA01001055 JAA93797.1 GDQN01005226 JAT85828.1 GECZ01003608 JAS66161.1 CH379064 EAL31971.1 GFTR01005427 JAW10999.1 GBGD01001916 JAC86973.1 KJ736824 CH478475 AIF75096.1 EAT32915.1 GAIX01003374 JAA89186.1 GALA01001056 JAA93796.1 CP012528 ALC48311.1 CVRI01000024 CRK92217.1 BT128214 AEE63175.1 GL435038 EFN74126.1 CH933811 EDW06324.1
NWSH01000131 PCG79222.1 GAIX01009046 JAA83514.1 BABH01015047 BABH01015048 BABH01015049 PCG79221.1 KQ460711 KPJ12677.1 KQ459603 KPI93114.1 GL448810 EFN83781.1 KQ982638 KYQ53297.1 KQ976445 KYM86057.1 GL888193 EGI65404.1 KQ980794 KYN13057.1 QOIP01000004 RLU23891.1 KQ981522 KYN40674.1 GL434491 EFN74876.1 KK107046 EZA61697.1 GAKP01016340 JAC42612.1 GBHO01044300 GBHO01030105 GBRD01012033 GDHC01002278 JAF99303.1 JAG13499.1 JAG53791.1 JAQ16351.1 GDHF01015923 GDHF01012857 JAI36391.1 JAI39457.1 GEDC01023218 GEDC01018802 JAS14080.1 JAS18496.1 NEVH01025142 PNF15718.1 KQ978143 KYM96758.1 JXJN01001234 EF197891 ABM88790.1 BABH01003247 AY461704 AAR97567.2 CCAG010000780 KQ459596 KPI95361.1 KQ460497 KPJ14319.1 GAMC01015097 JAB91458.1 GAMC01015098 JAB91457.1 GALX01004058 JAB64408.1 AJVK01014295 AXCM01003927 PYGN01000272 PSN49595.1 GBXI01002190 JAD12102.1 AE014298 ACZ95220.1 AF137269 AF172257 AY060368 AY196138 ODYU01003264 AAP40732.1 SOQ41789.1 CH954180 EDV47434.1 AY570724 AAS77384.1 GFDF01000920 JAV13164.1 KK853092 KDR11527.1 KA646715 AFP61344.1 AJWK01030336 CM000162 EDX02415.1 CH940655 EDW66021.1 AF115854 FJ619503 ACN41954.1 AXCN02001524 JRES01001325 KNC23628.1 KC018471 AFY62975.1 KC018473 AFY62977.1 NWSH01000281 PCG77801.1 KK855997 PTY24583.1 GEBQ01022017 JAT17960.1 CH480839 EDW49337.1 ATLV01019742 KE525279 KFB44592.1 ADTU01026241 CH902621 EDV44437.1 GANO01001288 JAB58583.1 RSAL01000001 RVE55224.1 CH964239 EDW82129.1 OUUW01000016 SPP88803.1 GDIQ01002289 JAN92448.1 LNIX01000003 OXA57277.1 CH916371 EDV92024.1 GECU01021896 JAS85810.1 AGBW02011483 OWR46576.1 JXUM01157673 KQ572819 KXJ67988.1 KQ971380 EEZ97563.1 GALA01001055 JAA93797.1 GDQN01005226 JAT85828.1 GECZ01003608 JAS66161.1 CH379064 EAL31971.1 GFTR01005427 JAW10999.1 GBGD01001916 JAC86973.1 KJ736824 CH478475 AIF75096.1 EAT32915.1 GAIX01003374 JAA89186.1 GALA01001056 JAA93796.1 CP012528 ALC48311.1 CVRI01000024 CRK92217.1 BT128214 AEE63175.1 GL435038 EFN74126.1 CH933811 EDW06324.1
Proteomes
UP000218220
UP000005204
UP000053240
UP000053268
UP000008237
UP000075809
+ More
UP000078540 UP000007755 UP000078492 UP000279307 UP000078541 UP000000311 UP000053097 UP000235965 UP000075881 UP000075920 UP000078542 UP000092443 UP000092460 UP000092444 UP000078200 UP000092445 UP000076408 UP000075884 UP000092462 UP000075883 UP000245037 UP000000803 UP000008711 UP000075900 UP000027135 UP000095301 UP000092461 UP000002282 UP000008792 UP000095300 UP000075886 UP000037069 UP000091820 UP000001292 UP000030765 UP000005205 UP000007801 UP000283053 UP000007798 UP000268350 UP000075901 UP000198287 UP000001070 UP000007151 UP000069940 UP000249989 UP000192223 UP000192221 UP000007266 UP000001819 UP000008820 UP000092553 UP000183832 UP000009192
UP000078540 UP000007755 UP000078492 UP000279307 UP000078541 UP000000311 UP000053097 UP000235965 UP000075881 UP000075920 UP000078542 UP000092443 UP000092460 UP000092444 UP000078200 UP000092445 UP000076408 UP000075884 UP000092462 UP000075883 UP000245037 UP000000803 UP000008711 UP000075900 UP000027135 UP000095301 UP000092461 UP000002282 UP000008792 UP000095300 UP000075886 UP000037069 UP000091820 UP000001292 UP000030765 UP000005205 UP000007801 UP000283053 UP000007798 UP000268350 UP000075901 UP000198287 UP000001070 UP000007151 UP000069940 UP000249989 UP000192223 UP000192221 UP000007266 UP000001819 UP000008820 UP000092553 UP000183832 UP000009192
Interpro
IPR000990
Innexin
+ More
IPR016169 FAD-bd_PCMH_sub2
IPR036318 FAD-bd_PCMH-like_sf
IPR016167 FAD-bd_PCMH_sub1
IPR004113 FAD-linked_oxidase_C
IPR025650 Alkyl-DHAP_Synthase
IPR016166 FAD-bd_PCMH
IPR016164 FAD-linked_Oxase-like_C
IPR016171 Vanillyl_alc_oxidase_C-sub2
IPR006094 Oxid_FAD_bind_N
IPR023123 Tubulin_C
IPR008280 Tub_FtsZ_C
IPR016169 FAD-bd_PCMH_sub2
IPR036318 FAD-bd_PCMH-like_sf
IPR016167 FAD-bd_PCMH_sub1
IPR004113 FAD-linked_oxidase_C
IPR025650 Alkyl-DHAP_Synthase
IPR016166 FAD-bd_PCMH
IPR016164 FAD-linked_Oxase-like_C
IPR016171 Vanillyl_alc_oxidase_C-sub2
IPR006094 Oxid_FAD_bind_N
IPR023123 Tubulin_C
IPR008280 Tub_FtsZ_C
Gene 3D
ProteinModelPortal
Q5XLD8
A2TEZ1
A0A2H1VAA9
A0A2A4K6C4
S4P161
H9JC80
+ More
A0A2A4K4P2 A0A0N1PHR1 A0A194PK59 E2BKJ4 A0A151WZW3 A0A151I4W9 F4WK32 A0A195DJR1 A0A3L8DU44 A0A195FL04 E1ZVA0 A0A026X0B1 A0A034VKH4 A0A0A9VUC1 A0A0K8VKP7 A0A1B6CKW9 A0A2J7PHC1 A0A182JZM3 A0A182VWE0 A0A195C9G7 A0A1A9XRV8 A0A1B0AP71 A2TEZ0 Q6SA04 A0A1B0G4W2 A0A1A9UUN2 A0A1A9ZE59 A0A182YB92 A0A182NA24 A0A194PPJ2 A0A0N0PCR1 W8B342 W8AZ10 V5GVY7 A0A1B0DD12 A0A182M026 A0A2P8YZB7 A0A0A1XMB6 E1JJF3 Q9V427 Q6XZI1 B3NXY3 Q6PYX4 A0A182R506 A0A1L8E3F2 A0A067R1S3 T1PEY5 A0A1B0CVA1 B4Q160 B4M9W2 Q9XYN1 A0A1I8P411 C0KLS6 A0A182QM11 A0A0L0BUG1 K9RY87 K9S0V4 A0A2A4K0K9 A0A2R7WYH0 A0A1A9WPK2 A0A1B6L2T6 B4IHI6 A0A084W2Z8 A0A158NU57 B3MQ11 U5EUV3 A0A3S2LJH0 B4NEG0 A0A3B0K691 A0A0P6IZ10 A0A182SMM5 A0A226EJI1 B4JMT1 A0A1B6IFW0 A0A212EYK5 A0A182GB47 A0A1W4WZU8 A0A1W4V8D0 D6X4J0 T1E2R6 A0A1E1WFT8 A0A1B6GUR0 Q29GY8 A0A224XSQ0 A0A069DSD1 Q16FA2 S4PA74 T1DI93 A0A0M5J2R8 A0A1J1I1G9 J3JY90 E1ZXE9 B4L565
A0A2A4K4P2 A0A0N1PHR1 A0A194PK59 E2BKJ4 A0A151WZW3 A0A151I4W9 F4WK32 A0A195DJR1 A0A3L8DU44 A0A195FL04 E1ZVA0 A0A026X0B1 A0A034VKH4 A0A0A9VUC1 A0A0K8VKP7 A0A1B6CKW9 A0A2J7PHC1 A0A182JZM3 A0A182VWE0 A0A195C9G7 A0A1A9XRV8 A0A1B0AP71 A2TEZ0 Q6SA04 A0A1B0G4W2 A0A1A9UUN2 A0A1A9ZE59 A0A182YB92 A0A182NA24 A0A194PPJ2 A0A0N0PCR1 W8B342 W8AZ10 V5GVY7 A0A1B0DD12 A0A182M026 A0A2P8YZB7 A0A0A1XMB6 E1JJF3 Q9V427 Q6XZI1 B3NXY3 Q6PYX4 A0A182R506 A0A1L8E3F2 A0A067R1S3 T1PEY5 A0A1B0CVA1 B4Q160 B4M9W2 Q9XYN1 A0A1I8P411 C0KLS6 A0A182QM11 A0A0L0BUG1 K9RY87 K9S0V4 A0A2A4K0K9 A0A2R7WYH0 A0A1A9WPK2 A0A1B6L2T6 B4IHI6 A0A084W2Z8 A0A158NU57 B3MQ11 U5EUV3 A0A3S2LJH0 B4NEG0 A0A3B0K691 A0A0P6IZ10 A0A182SMM5 A0A226EJI1 B4JMT1 A0A1B6IFW0 A0A212EYK5 A0A182GB47 A0A1W4WZU8 A0A1W4V8D0 D6X4J0 T1E2R6 A0A1E1WFT8 A0A1B6GUR0 Q29GY8 A0A224XSQ0 A0A069DSD1 Q16FA2 S4PA74 T1DI93 A0A0M5J2R8 A0A1J1I1G9 J3JY90 E1ZXE9 B4L565
PDB
5H1R
E-value=6.49742e-11,
Score=162
Ontologies
GO
GO:0005921
GO:0006811
GO:0005886
GO:0016021
GO:0016491
GO:0008610
GO:0071949
GO:0008609
GO:0005243
GO:0016324
GO:0005737
GO:0007293
GO:0007154
GO:0016331
GO:0007440
GO:0007283
GO:0010496
GO:0016328
GO:0016323
GO:0030727
GO:0016327
GO:0036098
GO:0003824
GO:0016614
GO:0005852
GO:0006413
GO:0031369
GO:0005515
GO:0046872
Topology
Subcellular location
Cell membrane
Cell junction
Gap junction
Cytoplasm
Apical cell membrane
Apicolateral cell membrane
Basolateral cell membrane
Lateral cell membrane
Cell junction
Gap junction
Cytoplasm
Apical cell membrane
Apicolateral cell membrane
Basolateral cell membrane
Lateral cell membrane
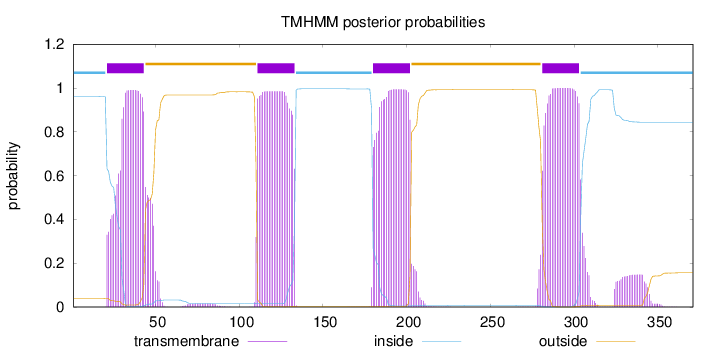
Length:
371
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
93.5126400000001
Exp number, first 60 AAs:
21.07015
Total prob of N-in:
0.96080
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 43
outside
44 - 110
TMhelix
111 - 133
inside
134 - 179
TMhelix
180 - 202
outside
203 - 280
TMhelix
281 - 303
inside
304 - 371
Population Genetic Test Statistics
Pi
232.798146
Theta
200.751791
Tajima's D
0.281875
CLR
2.105894
CSRT
0.452477376131193
Interpretation
Uncertain
