Gene
KWMTBOMO10515 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007125
Annotation
PREDICTED:_alkyldihydroxyacetonephosphate_synthase_[Amyelois_transitella]
Full name
Innexin
+ More
Alkylglycerone-phosphate synthase
Alkyldihydroxyacetonephosphate synthase
Alkylglycerone-phosphate synthase
Alkyldihydroxyacetonephosphate synthase
Alternative Name
Alkylglycerone-phosphate synthase
Location in the cell
Cytoplasmic Reliability : 1.927
Sequence
CDS
ATGAACTGGCCATTTTTCCTTGGACAGATATTGTACCTTGTAAGAGCAGAGAATATGCTCTCCGACGAGGCTGCATTGTTTGTTGCTTATGTGCACGATAGTGGTCTTGATCGTCATGTAGTGTTGAAAGGGGCAGACTTGATGCGTCTCACGATGTCGCTCCATCTCTCCCTGCTCGAGGCCATGTCATCACAATACAATTTAAGACGTTCACCAAGCATCTTTTGGTGTAGTCGATACTCAATAGCAGGGAAATGCTTACCCCACCTACCGCAGTGGGCCATAGACAACTTTGGCATAGATCCAGATTCCCCGCCCAAAATTCCGGAAGCGCCAAGTAGTTTTGCAGAAAGTCGCCTGCCGGAAAACGTTAAGGATGAATTGGAGATGATCGCCGCAGTAAGCGTGGACGGCATGGATAGACTGATCAGAGCACACGGGCAGACCTTAAAGGACATAGCGAACGTCAGAAATAACAACTTTCGGAGAATACCTGACGCTGTCATATGGCCAGATTGTCATGAGCAGGTAGAAAAGATAGTCCAATGTGCGTCGCGGCACAACTTCGTGATCATTCCTTTCGGTGGGGGCACGTCGGTGTCCGGCGCGGTCACTTGCCCCGAGAACGAACCCAGGCCCATAGTCGCCCTGGACACGAGCGAAATGAATTCAATACTGTGGTTGGACAAGGAGCAGCTGCTGGCGAGAGTGCAAGCAGGAATAGTCGGTCAAGACTTAGAGCGGGAGATGCGGTCCCGTGGCTTCACCGTCGGCCACGAGCCAGACTCGTACGAGTTCTCTACCCTCGGGGGGTGGGTTGCGACAAGAGCCAGCGGCATGAAGAAGAACACATATGGGAATATAGAAGATTTAGTTGTACAGACGAAGGTGGTGACGCCGAAAGGAGTGCTCGAGAAGAGCTGTCGCGTGCCGAGGGTGTCGTGCGGTCCGGAATGGGAGCACGTCGTGATGGGCTCCGAAGGATGCTTCGGTGTCGTCACTGAGGTCGTCATCAAAATCCGTCCAGTGCCGGCCGTGACTCGCTACGGATCCCTCGTGTTCCCCGACTGGGAGTCCGGGTTCTGGTTCGAAAGGGAAGTCGCCAGGAAGAGGCTTCAACCTTCTAGTATTAGACTAATGGACAACGAACAGTTTCGTTTCGGGCAAGCACTGAAATTGGAGAACTCGTGGGGCGGAGTCCTGTTGGACGGGCTGAAGCGACTGTACATCACTAGGATCAAAGGGTTCGATCCTCTGAAGCTGTGCGTGGTCACGCTGCTCATGGAGGGCACCGAGGAGAAAGTCAGAGAACAAGAGCGAGCGCTCAACGGCATCGCGGCCCGGCACGGCGGGGTGCCGGCCGGCGCGGGGAACGGCGAGACCGGATACACGCTCACCTTCGTCATAGCCTACCTCAGGGACGTGGCGATAGAGTTCGACATAGTGGCGGAATCGTTCGAGACGTCGGTGCCCTGGGACCGCACCCTGGCTCTCTGCAGGAACGTGAAGACTCGCGTCCGGGACGAGTGTCTCGCCAGAAGTATCACCCGGTTCCTGATCTCGTGCCGCCTCACTCAAACTTACGACGCCGGCTGCTGTATTTATTTTTATTTTGGATTTTACTCGGGTGACTGTGAAAACCCTGTCGAAACGTACGAGCAAATCGAGGAAGCAGCCAGAGATGAAATATTAGCTTGCGGTGGGTCGATATCGCACCATCATGGCGTCGGAAAATTAAGAAAGAAATGGTACGCCGAGACTGTGAGCGAACCCGGCCGCCAATTACTACTGGCAGCTAAGAACGCCTTAGATCCGCAGAACATATTTGCAGTCGGCAATATGGCCTTTGACGAGCACAACACCGAACATAGCGTCAAAAGTAAACTATGA
Protein
MNWPFFLGQILYLVRAENMLSDEAALFVAYVHDSGLDRHVVLKGADLMRLTMSLHLSLLEAMSSQYNLRRSPSIFWCSRYSIAGKCLPHLPQWAIDNFGIDPDSPPKIPEAPSSFAESRLPENVKDELEMIAAVSVDGMDRLIRAHGQTLKDIANVRNNNFRRIPDAVIWPDCHEQVEKIVQCASRHNFVIIPFGGGTSVSGAVTCPENEPRPIVALDTSEMNSILWLDKEQLLARVQAGIVGQDLEREMRSRGFTVGHEPDSYEFSTLGGWVATRASGMKKNTYGNIEDLVVQTKVVTPKGVLEKSCRVPRVSCGPEWEHVVMGSEGCFGVVTEVVIKIRPVPAVTRYGSLVFPDWESGFWFEREVARKRLQPSSIRLMDNEQFRFGQALKLENSWGGVLLDGLKRLYITRIKGFDPLKLCVVTLLMEGTEEKVREQERALNGIAARHGGVPAGAGNGETGYTLTFVIAYLRDVAIEFDIVAESFETSVPWDRTLALCRNVKTRVRDECLARSITRFLISCRLTQTYDAGCCIYFYFGFYSGDCENPVETYEQIEEAARDEILACGGSISHHHGVGKLRKKWYAETVSEPGRQLLLAAKNALDPQNIFAVGNMAFDEHNTEHSVKSKL
Summary
Description
Structural component of the gap junctions.
Catalyzes the exchange of an acyl for a long-chain alkyl group and the formation of the ether bond in the biosynthesis of ether phospholipids.
Catalyzes the exchange of an acyl for a long-chain alkyl group and the formation of the ether bond in the biosynthesis of ether phospholipids.
Catalytic Activity
a 1-acylglycerone 3-phosphate + a long chain fatty alcohol = 1-O-alkylglycerone 3-phosphate + a long-chain fatty acid + H(+)
Cofactor
FAD
Subunit
Homodimer.
Similarity
Belongs to the pannexin family.
Belongs to the FAD-binding oxidoreductase/transferase type 4 family.
Belongs to the FAD-binding oxidoreductase/transferase type 4 family.
Keywords
Complete proteome
FAD
Flavoprotein
Lipid biosynthesis
Lipid metabolism
Peroxisome
Reference proteome
Transferase
Feature
chain Innexin
Uniprot
H9JC80
A0A2H1VA52
A0A2A4K6C2
A0A194PJ97
D6WW52
A0A1B0C9B7
+ More
A0A2J7Q507 A0A182K051 A0A182Q4S0 A0A182VPM3 A0A1B6IX74 A0A182P3E9 A0A182YJQ0 A0A1B6EHV6 A0A1L8DAN0 A0A1L8DAN4 A0A034VKX9 A0A182R8H3 J3JW52 A0A1L8DAZ9 A0A2P8YUN1 A0A182SQW2 A0A0K8VS84 N6TXY9 A0A182LV29 A0A182LNJ1 A0A1B6LVZ4 A0A0M4EHA8 Q7QA93 A0A182XF27 A0A182UXZ6 A0A182HVJ5 A0A182NCQ5 W5JEK9 B3MH07 A0A182FPJ7 A0A0B4JD21 Q9V778 B3NQQ2 B4HRS2 A0A3B0JS51 B4P7R8 A0A1I8MB57 A0A1A9VZE3 A0A1B0GBE9 B4QGC0 D5SHU3 A0A1A9ZPI4 E0VIW8 A0A182TZH2 A0A0A1WK86 T1PK60 A0A1W4UM17 A0A1Y1MVA3 A0A1Y1MS72 A0A1B0B4G9 K7JA90 B4LLK6 W8BYB7 A0A1A9VJD3 A0A1B6CGH0 B4KP39 A0A182JL87 B5E051 A0A1Z5KWP2 A0A0L0CMS2 A0A336N0B4 B4GD67 A0A2A3E405 V9IGF4 A0A310SLM8 A0A336MYR2 A0A151X1B3 B4J5Y8 B4MQ03 A0A1I8P8Q1 A0A1Q3FIE7 A0A224YJ86 B0WMX1 A0A131Z8X7 A0A1D2NHA7 T1H9K7 A0A0N8ES99 A0A2R5LLX7 A0A154P064 A0A151I0V8 A0A146L266 A0A151I8K5 A0A1E1X931 A0A158NAD8 A0A026WRM0 A0A210QBG3 A0A195EW99 A0A1Y1MTG5 F4WMI0 A0A069DWA1 A0A023F3H5 A0A151LZZ3 A0A1U8DJK3 A0A1S3JUX1
A0A2J7Q507 A0A182K051 A0A182Q4S0 A0A182VPM3 A0A1B6IX74 A0A182P3E9 A0A182YJQ0 A0A1B6EHV6 A0A1L8DAN0 A0A1L8DAN4 A0A034VKX9 A0A182R8H3 J3JW52 A0A1L8DAZ9 A0A2P8YUN1 A0A182SQW2 A0A0K8VS84 N6TXY9 A0A182LV29 A0A182LNJ1 A0A1B6LVZ4 A0A0M4EHA8 Q7QA93 A0A182XF27 A0A182UXZ6 A0A182HVJ5 A0A182NCQ5 W5JEK9 B3MH07 A0A182FPJ7 A0A0B4JD21 Q9V778 B3NQQ2 B4HRS2 A0A3B0JS51 B4P7R8 A0A1I8MB57 A0A1A9VZE3 A0A1B0GBE9 B4QGC0 D5SHU3 A0A1A9ZPI4 E0VIW8 A0A182TZH2 A0A0A1WK86 T1PK60 A0A1W4UM17 A0A1Y1MVA3 A0A1Y1MS72 A0A1B0B4G9 K7JA90 B4LLK6 W8BYB7 A0A1A9VJD3 A0A1B6CGH0 B4KP39 A0A182JL87 B5E051 A0A1Z5KWP2 A0A0L0CMS2 A0A336N0B4 B4GD67 A0A2A3E405 V9IGF4 A0A310SLM8 A0A336MYR2 A0A151X1B3 B4J5Y8 B4MQ03 A0A1I8P8Q1 A0A1Q3FIE7 A0A224YJ86 B0WMX1 A0A131Z8X7 A0A1D2NHA7 T1H9K7 A0A0N8ES99 A0A2R5LLX7 A0A154P064 A0A151I0V8 A0A146L266 A0A151I8K5 A0A1E1X931 A0A158NAD8 A0A026WRM0 A0A210QBG3 A0A195EW99 A0A1Y1MTG5 F4WMI0 A0A069DWA1 A0A023F3H5 A0A151LZZ3 A0A1U8DJK3 A0A1S3JUX1
EC Number
2.5.1.26
Pubmed
19121390
26354079
18362917
19820115
25244985
25348373
+ More
22516182 23537049 29403074 20966253 12364791 14747013 17210077 20920257 23761445 17994087 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12537569 17550304 25315136 22936249 20566863 25830018 28004739 20075255 24495485 15632085 23185243 28528879 26108605 28797301 26830274 27289101 26999592 26823975 28503490 21347285 24508170 30249741 28812685 21719571 26334808 25474469 22293439
22516182 23537049 29403074 20966253 12364791 14747013 17210077 20920257 23761445 17994087 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12537569 17550304 25315136 22936249 20566863 25830018 28004739 20075255 24495485 15632085 23185243 28528879 26108605 28797301 26830274 27289101 26999592 26823975 28503490 21347285 24508170 30249741 28812685 21719571 26334808 25474469 22293439
EMBL
BABH01015047
BABH01015048
BABH01015049
ODYU01001480
SOQ37730.1
NWSH01000131
+ More
PCG79220.1 KQ459603 KPI93113.1 KQ971361 EFA08666.1 AJWK01002156 NEVH01018372 PNF23665.1 AXCN02000334 GECU01016171 JAS91535.1 GECZ01032261 JAS37508.1 GFDF01010566 JAV03518.1 GFDF01010565 JAV03519.1 GAKP01016472 JAC42480.1 BT127470 KB631786 AEE62432.1 ERL86064.1 GFDF01010564 JAV03520.1 PYGN01000350 PSN47945.1 GDHF01010585 JAI41729.1 APGK01056813 KB741277 ENN71142.1 AXCM01000757 AXCM01000758 AXCM01000759 GEBQ01019593 GEBQ01012700 GEBQ01012139 JAT20384.1 JAT27277.1 JAT27838.1 CP012524 ALC41922.1 AAAB01008898 EAA09140.2 APCN01005786 APCN01005787 ADMH02001712 ETN61330.1 CH902619 EDV35766.1 KPU75815.1 AE013599 ADV37181.1 AY094917 CH954179 EDV55962.1 CH480816 EDW47936.1 OUUW01000001 SPP75531.1 CM000158 EDW91094.1 CCAG010013992 CM000362 CM002911 EDX07158.1 KMY93874.1 BT124894 ADG57801.1 DS235212 EEB13324.1 GBXI01015196 JAC99095.1 KA648348 AFP62977.1 GEZM01023333 JAV88460.1 GEZM01023334 JAV88459.1 JXJN01008310 AAZX01000485 CH940648 EDW60869.1 KRF79646.1 GAMC01012301 JAB94254.1 GEDC01024809 JAS12489.1 CH933808 EDW10105.1 CM000071 EDY69284.1 KRT02782.1 GFJQ02007515 JAV99454.1 JRES01000171 KNC33648.1 UFQT01002967 SSX34373.1 CH479181 EDW32562.1 KZ288419 PBC25986.1 JR041348 AEY59419.1 KQ759894 OAD62044.1 UFQS01002628 UFQT01002628 SSX14393.1 SSX33803.1 KQ982585 KYQ54192.1 CH916367 EDW00831.1 CH963849 EDW74192.1 GFDL01007708 JAV27337.1 GFPF01005879 MAA17025.1 DS232003 EDS31363.1 GEDV01001139 JAP87418.1 LJIJ01000039 ODN04640.1 ACPB03024436 ACPB03024437 GDUN01000058 JAN95861.1 GGLE01006398 MBY10524.1 KQ434782 KZC04648.1 KQ976600 KYM79485.1 GDHC01016794 JAQ01835.1 KQ978355 KYM94631.1 GFAC01003429 JAT95759.1 ADTU01010258 ADTU01010259 ADTU01010260 KK107119 QOIP01000009 EZA58662.1 RLU18282.1 NEDP02004288 OWF46074.1 KQ981954 KYN32164.1 GEZM01023335 JAV88458.1 GL888217 EGI64591.1 GBGD01000937 JAC87952.1 GBBI01002814 JAC15898.1 AKHW03006853 KYO17790.1
PCG79220.1 KQ459603 KPI93113.1 KQ971361 EFA08666.1 AJWK01002156 NEVH01018372 PNF23665.1 AXCN02000334 GECU01016171 JAS91535.1 GECZ01032261 JAS37508.1 GFDF01010566 JAV03518.1 GFDF01010565 JAV03519.1 GAKP01016472 JAC42480.1 BT127470 KB631786 AEE62432.1 ERL86064.1 GFDF01010564 JAV03520.1 PYGN01000350 PSN47945.1 GDHF01010585 JAI41729.1 APGK01056813 KB741277 ENN71142.1 AXCM01000757 AXCM01000758 AXCM01000759 GEBQ01019593 GEBQ01012700 GEBQ01012139 JAT20384.1 JAT27277.1 JAT27838.1 CP012524 ALC41922.1 AAAB01008898 EAA09140.2 APCN01005786 APCN01005787 ADMH02001712 ETN61330.1 CH902619 EDV35766.1 KPU75815.1 AE013599 ADV37181.1 AY094917 CH954179 EDV55962.1 CH480816 EDW47936.1 OUUW01000001 SPP75531.1 CM000158 EDW91094.1 CCAG010013992 CM000362 CM002911 EDX07158.1 KMY93874.1 BT124894 ADG57801.1 DS235212 EEB13324.1 GBXI01015196 JAC99095.1 KA648348 AFP62977.1 GEZM01023333 JAV88460.1 GEZM01023334 JAV88459.1 JXJN01008310 AAZX01000485 CH940648 EDW60869.1 KRF79646.1 GAMC01012301 JAB94254.1 GEDC01024809 JAS12489.1 CH933808 EDW10105.1 CM000071 EDY69284.1 KRT02782.1 GFJQ02007515 JAV99454.1 JRES01000171 KNC33648.1 UFQT01002967 SSX34373.1 CH479181 EDW32562.1 KZ288419 PBC25986.1 JR041348 AEY59419.1 KQ759894 OAD62044.1 UFQS01002628 UFQT01002628 SSX14393.1 SSX33803.1 KQ982585 KYQ54192.1 CH916367 EDW00831.1 CH963849 EDW74192.1 GFDL01007708 JAV27337.1 GFPF01005879 MAA17025.1 DS232003 EDS31363.1 GEDV01001139 JAP87418.1 LJIJ01000039 ODN04640.1 ACPB03024436 ACPB03024437 GDUN01000058 JAN95861.1 GGLE01006398 MBY10524.1 KQ434782 KZC04648.1 KQ976600 KYM79485.1 GDHC01016794 JAQ01835.1 KQ978355 KYM94631.1 GFAC01003429 JAT95759.1 ADTU01010258 ADTU01010259 ADTU01010260 KK107119 QOIP01000009 EZA58662.1 RLU18282.1 NEDP02004288 OWF46074.1 KQ981954 KYN32164.1 GEZM01023335 JAV88458.1 GL888217 EGI64591.1 GBGD01000937 JAC87952.1 GBBI01002814 JAC15898.1 AKHW03006853 KYO17790.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007266
UP000092461
UP000235965
+ More
UP000075881 UP000075886 UP000075920 UP000075885 UP000076408 UP000075900 UP000030742 UP000245037 UP000075901 UP000019118 UP000075883 UP000075882 UP000092553 UP000007062 UP000076407 UP000075903 UP000075840 UP000075884 UP000000673 UP000007801 UP000069272 UP000000803 UP000008711 UP000001292 UP000268350 UP000002282 UP000095301 UP000091820 UP000092444 UP000000304 UP000092445 UP000009046 UP000075902 UP000192221 UP000092460 UP000002358 UP000008792 UP000078200 UP000009192 UP000075880 UP000001819 UP000037069 UP000008744 UP000242457 UP000075809 UP000001070 UP000007798 UP000095300 UP000002320 UP000094527 UP000015103 UP000076502 UP000078540 UP000078542 UP000005205 UP000053097 UP000279307 UP000242188 UP000078541 UP000007755 UP000050525 UP000189705 UP000085678
UP000075881 UP000075886 UP000075920 UP000075885 UP000076408 UP000075900 UP000030742 UP000245037 UP000075901 UP000019118 UP000075883 UP000075882 UP000092553 UP000007062 UP000076407 UP000075903 UP000075840 UP000075884 UP000000673 UP000007801 UP000069272 UP000000803 UP000008711 UP000001292 UP000268350 UP000002282 UP000095301 UP000091820 UP000092444 UP000000304 UP000092445 UP000009046 UP000075902 UP000192221 UP000092460 UP000002358 UP000008792 UP000078200 UP000009192 UP000075880 UP000001819 UP000037069 UP000008744 UP000242457 UP000075809 UP000001070 UP000007798 UP000095300 UP000002320 UP000094527 UP000015103 UP000076502 UP000078540 UP000078542 UP000005205 UP000053097 UP000279307 UP000242188 UP000078541 UP000007755 UP000050525 UP000189705 UP000085678
Interpro
IPR016169
FAD-bd_PCMH_sub2
+ More
IPR036318 FAD-bd_PCMH-like_sf
IPR016167 FAD-bd_PCMH_sub1
IPR004113 FAD-linked_oxidase_C
IPR025650 Alkyl-DHAP_Synthase
IPR016166 FAD-bd_PCMH
IPR016164 FAD-linked_Oxase-like_C
IPR000990 Innexin
IPR016171 Vanillyl_alc_oxidase_C-sub2
IPR006094 Oxid_FAD_bind_N
IPR019786 Zinc_finger_PHD-type_CS
IPR039054 Int12_PHD
IPR011011 Znf_FYVE_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR019787 Znf_PHD-finger
IPR036318 FAD-bd_PCMH-like_sf
IPR016167 FAD-bd_PCMH_sub1
IPR004113 FAD-linked_oxidase_C
IPR025650 Alkyl-DHAP_Synthase
IPR016166 FAD-bd_PCMH
IPR016164 FAD-linked_Oxase-like_C
IPR000990 Innexin
IPR016171 Vanillyl_alc_oxidase_C-sub2
IPR006094 Oxid_FAD_bind_N
IPR019786 Zinc_finger_PHD-type_CS
IPR039054 Int12_PHD
IPR011011 Znf_FYVE_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR019787 Znf_PHD-finger
Gene 3D
ProteinModelPortal
H9JC80
A0A2H1VA52
A0A2A4K6C2
A0A194PJ97
D6WW52
A0A1B0C9B7
+ More
A0A2J7Q507 A0A182K051 A0A182Q4S0 A0A182VPM3 A0A1B6IX74 A0A182P3E9 A0A182YJQ0 A0A1B6EHV6 A0A1L8DAN0 A0A1L8DAN4 A0A034VKX9 A0A182R8H3 J3JW52 A0A1L8DAZ9 A0A2P8YUN1 A0A182SQW2 A0A0K8VS84 N6TXY9 A0A182LV29 A0A182LNJ1 A0A1B6LVZ4 A0A0M4EHA8 Q7QA93 A0A182XF27 A0A182UXZ6 A0A182HVJ5 A0A182NCQ5 W5JEK9 B3MH07 A0A182FPJ7 A0A0B4JD21 Q9V778 B3NQQ2 B4HRS2 A0A3B0JS51 B4P7R8 A0A1I8MB57 A0A1A9VZE3 A0A1B0GBE9 B4QGC0 D5SHU3 A0A1A9ZPI4 E0VIW8 A0A182TZH2 A0A0A1WK86 T1PK60 A0A1W4UM17 A0A1Y1MVA3 A0A1Y1MS72 A0A1B0B4G9 K7JA90 B4LLK6 W8BYB7 A0A1A9VJD3 A0A1B6CGH0 B4KP39 A0A182JL87 B5E051 A0A1Z5KWP2 A0A0L0CMS2 A0A336N0B4 B4GD67 A0A2A3E405 V9IGF4 A0A310SLM8 A0A336MYR2 A0A151X1B3 B4J5Y8 B4MQ03 A0A1I8P8Q1 A0A1Q3FIE7 A0A224YJ86 B0WMX1 A0A131Z8X7 A0A1D2NHA7 T1H9K7 A0A0N8ES99 A0A2R5LLX7 A0A154P064 A0A151I0V8 A0A146L266 A0A151I8K5 A0A1E1X931 A0A158NAD8 A0A026WRM0 A0A210QBG3 A0A195EW99 A0A1Y1MTG5 F4WMI0 A0A069DWA1 A0A023F3H5 A0A151LZZ3 A0A1U8DJK3 A0A1S3JUX1
A0A2J7Q507 A0A182K051 A0A182Q4S0 A0A182VPM3 A0A1B6IX74 A0A182P3E9 A0A182YJQ0 A0A1B6EHV6 A0A1L8DAN0 A0A1L8DAN4 A0A034VKX9 A0A182R8H3 J3JW52 A0A1L8DAZ9 A0A2P8YUN1 A0A182SQW2 A0A0K8VS84 N6TXY9 A0A182LV29 A0A182LNJ1 A0A1B6LVZ4 A0A0M4EHA8 Q7QA93 A0A182XF27 A0A182UXZ6 A0A182HVJ5 A0A182NCQ5 W5JEK9 B3MH07 A0A182FPJ7 A0A0B4JD21 Q9V778 B3NQQ2 B4HRS2 A0A3B0JS51 B4P7R8 A0A1I8MB57 A0A1A9VZE3 A0A1B0GBE9 B4QGC0 D5SHU3 A0A1A9ZPI4 E0VIW8 A0A182TZH2 A0A0A1WK86 T1PK60 A0A1W4UM17 A0A1Y1MVA3 A0A1Y1MS72 A0A1B0B4G9 K7JA90 B4LLK6 W8BYB7 A0A1A9VJD3 A0A1B6CGH0 B4KP39 A0A182JL87 B5E051 A0A1Z5KWP2 A0A0L0CMS2 A0A336N0B4 B4GD67 A0A2A3E405 V9IGF4 A0A310SLM8 A0A336MYR2 A0A151X1B3 B4J5Y8 B4MQ03 A0A1I8P8Q1 A0A1Q3FIE7 A0A224YJ86 B0WMX1 A0A131Z8X7 A0A1D2NHA7 T1H9K7 A0A0N8ES99 A0A2R5LLX7 A0A154P064 A0A151I0V8 A0A146L266 A0A151I8K5 A0A1E1X931 A0A158NAD8 A0A026WRM0 A0A210QBG3 A0A195EW99 A0A1Y1MTG5 F4WMI0 A0A069DWA1 A0A023F3H5 A0A151LZZ3 A0A1U8DJK3 A0A1S3JUX1
PDB
6GOU
E-value=2.09623e-172,
Score=1557
Ontologies
PATHWAY
GO
PANTHER
Topology
Subcellular location
Cell membrane
Cell junction
Gap junction
Peroxisome
Cell junction
Gap junction
Peroxisome
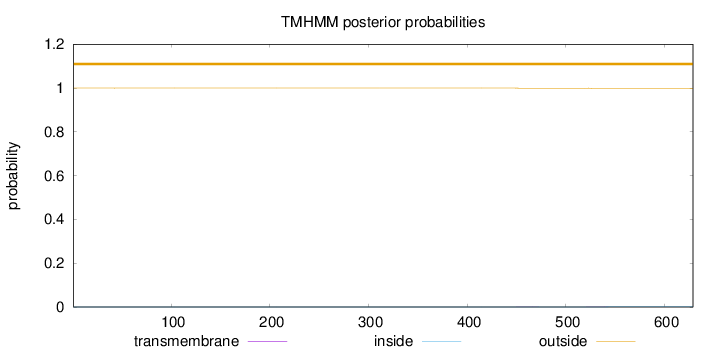
Length:
629
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02736
Exp number, first 60 AAs:
0.00113
Total prob of N-in:
0.00020
outside
1 - 629
Population Genetic Test Statistics
Pi
237.879774
Theta
172.220107
Tajima's D
0.102973
CLR
0.056973
CSRT
0.396830158492075
Interpretation
Uncertain
