Gene
KWMTBOMO10512
Pre Gene Modal
BGIBMGA007124
Annotation
PREDICTED:_uncharacterized_protein_LOC106142705_[Amyelois_transitella]
Location in the cell
Extracellular Reliability : 1.92 Nuclear Reliability : 1.895
Sequence
CDS
ATGTACTGCCTGCTAACTTCATTATTAGCAGTCCTTCTCGCAGCGGTAACAGCCTCCATCGGAGCTCCAGCATCATCAGCTAACTATGATGAGAACCTTACATTGGACAGAAAAGAGCAATCCACTCATGAATTATTAAGCTCGCTTGGCTTAAAAAAACTGAGAATACCGGCCGCGCATCACAGGAAGCGCCTCGCAGAACAAGGGAGACGCCATAGTACGGGAGACTCCAGAATGTACGTCATTAAACTACCCCCCAACATGAGTTATTACTCTCACGCTGATCCAGCGCCCGCCGCAGCGAGGACTCTACCTCTTCAGATGAAGAGCAACGGAAAACCTGCACGCGTTTACCACTGGAATATTCCAGTACTGCAGAAATTGGCAAAGAACCGACCTCAGGCAAGATTCGACGACGATAACATAGTCGATGTTGAAAGTACTCCAACGTGGCCAAAAGCACCGACCGATCATCCGAACTCTCTTGAAGCGTTAGCGTATTACGTTCCCGCCAAAAAGACATCGTTCCGAAAATATTTTTCAGGCAACGGGAAACCGAAATCATTTTATGTGATCGAAAAAAACAAGAGTGCCGCACAGTATCATAAGTTATTGCCTTAA
Protein
MYCLLTSLLAVLLAAVTASIGAPASSANYDENLTLDRKEQSTHELLSSLGLKKLRIPAAHHRKRLAEQGRRHSTGDSRMYVIKLPPNMSYYSHADPAPAAARTLPLQMKSNGKPARVYHWNIPVLQKLAKNRPQARFDDDNIVDVESTPTWPKAPTDHPNSLEALAYYVPAKKTSFRKYFSGNGKPKSFYVIEKNKSAAQYHKLLP
Summary
Uniprot
H9JC79
A0A0N1IGS6
I4DK90
A0A2H1VA69
A0A2A4K5B7
A0A0L7KMD6
+ More
A0A212F855 A0A1Y1KKL0 A0A1W4XA86 D6WVH8 N6TPB6 A0A1B6JT32 A0A2J7QAD6 A0A336M7X7 Q16U16 A0A2P8ZC83 A0A1B6FP74 A0A069DQW7 A0A1A9VJ25 A0A182G2Y5 A0A0A9WN19 A0A1B0CHW5 T1HLP8 A0A1A9Z7B4 W8AGT2 B4QY31 A0A1J1HPY7 A0A0A1X9P2 A0A1B6CNH4 A0A0K8VK24 A0A034VBJ7 A0A182QGK7 A0A0L0BQ79 A0A182R666 A0A182JWY5 A0A182P571 A0A182YAP4 A0A182MDE4 A0A182VZ74 A0A1B0FP45 A0A0M5J4N2 A0A182X0A8 A0A182HZZ8 A0A182N431 A0A1I8NB81 A0A182LR29 Q380S4 A0A182U967 B4M618 B4IGV1 A0A182VLE4 Q9VB07 A0A182JF85 A0A1I8Q6C0 A0A1I8MK34 A0A2R7WNT5 A0A182SLY3 B3LX33 A0A182F968 B4KAS7 B4JU62 B4NAR9 A0A1W4V8X7 B4GNX0 B3P5Y4 Q29BG2 A0A1A9Y2J8 B0X1Z1 A0A3B0JF15 E0VSP5 A0A2S2R2C5 J9KA21 A0A2S2NRH9 A0A0Q9XDR7 A0A154PQV0 A0A158NU45 E2BLF3 A0A151I557
A0A212F855 A0A1Y1KKL0 A0A1W4XA86 D6WVH8 N6TPB6 A0A1B6JT32 A0A2J7QAD6 A0A336M7X7 Q16U16 A0A2P8ZC83 A0A1B6FP74 A0A069DQW7 A0A1A9VJ25 A0A182G2Y5 A0A0A9WN19 A0A1B0CHW5 T1HLP8 A0A1A9Z7B4 W8AGT2 B4QY31 A0A1J1HPY7 A0A0A1X9P2 A0A1B6CNH4 A0A0K8VK24 A0A034VBJ7 A0A182QGK7 A0A0L0BQ79 A0A182R666 A0A182JWY5 A0A182P571 A0A182YAP4 A0A182MDE4 A0A182VZ74 A0A1B0FP45 A0A0M5J4N2 A0A182X0A8 A0A182HZZ8 A0A182N431 A0A1I8NB81 A0A182LR29 Q380S4 A0A182U967 B4M618 B4IGV1 A0A182VLE4 Q9VB07 A0A182JF85 A0A1I8Q6C0 A0A1I8MK34 A0A2R7WNT5 A0A182SLY3 B3LX33 A0A182F968 B4KAS7 B4JU62 B4NAR9 A0A1W4V8X7 B4GNX0 B3P5Y4 Q29BG2 A0A1A9Y2J8 B0X1Z1 A0A3B0JF15 E0VSP5 A0A2S2R2C5 J9KA21 A0A2S2NRH9 A0A0Q9XDR7 A0A154PQV0 A0A158NU45 E2BLF3 A0A151I557
Pubmed
19121390
26354079
22651552
26227816
22118469
28004739
+ More
18362917 19820115 23537049 17510324 29403074 26334808 26483478 25401762 26823975 24495485 17994087 25830018 25348373 26108605 25244985 25315136 20966253 12364791 14747013 17210077 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 20566863 18057021 21347285 20798317
18362917 19820115 23537049 17510324 29403074 26334808 26483478 25401762 26823975 24495485 17994087 25830018 25348373 26108605 25244985 25315136 20966253 12364791 14747013 17210077 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 20566863 18057021 21347285 20798317
EMBL
BABH01015044
KQ460711
KPJ12674.1
AK401708
BAM18330.1
ODYU01001480
+ More
SOQ37729.1 NWSH01000131 PCG79216.1 JTDY01008990 KOB64231.1 AGBW02009792 OWR49916.1 GEZM01083215 JAV61148.1 KQ971357 EFA08569.2 APGK01057182 KB741278 KB632283 ENN71060.1 ERL91395.1 GECU01005326 JAT02381.1 NEVH01016331 PNF25532.1 UFQT01000234 SSX22148.1 CH477634 EAT38026.1 PYGN01000107 PSN54116.1 GECZ01017810 JAS51959.1 GBGD01002788 JAC86101.1 JXUM01140342 JXUM01140343 JXUM01140344 JXUM01140345 KQ569047 KXJ68784.1 GBHO01037354 GBRD01013668 GDHC01012314 JAG06250.1 JAG52158.1 JAQ06315.1 AJWK01012785 ACPB03001521 GAMC01021703 JAB84852.1 CM000364 EDX14679.1 CVRI01000008 CRK88534.1 GBXI01006914 JAD07378.1 GEDC01022360 GEDC01021910 JAS14938.1 JAS15388.1 GDHF01023585 GDHF01013384 JAI28729.1 JAI38930.1 GAKP01020027 GAKP01020024 JAC38928.1 AXCN02000023 JRES01001623 KNC21364.1 AXCM01003710 AXCM01003711 CCAG010023115 CP012526 ALC45968.1 APCN01002026 AAAB01008987 EAL38486.3 CH940652 EDW59094.2 CH480837 EDW49069.1 AE014297 AY061370 AAF56738.2 AAL28918.1 KK855175 PTY21286.1 CH902617 EDV43872.1 CH933806 EDW16814.2 CH916374 EDV91032.1 CH964232 EDW80883.1 CH479186 EDW38853.1 CH954182 EDV53384.1 CM000070 EAL27036.3 DS232275 EDS38921.1 OUUW01000005 SPP80715.1 DS235755 EEB16401.1 GGMS01014988 MBY84191.1 ABLF02031876 ABLF02031877 GGMR01007136 MBY19755.1 KRG02311.1 KRG02312.1 KQ435010 KZC13724.1 ADTU01026200 GL449017 EFN83505.1 KQ976445 KYM86099.1
SOQ37729.1 NWSH01000131 PCG79216.1 JTDY01008990 KOB64231.1 AGBW02009792 OWR49916.1 GEZM01083215 JAV61148.1 KQ971357 EFA08569.2 APGK01057182 KB741278 KB632283 ENN71060.1 ERL91395.1 GECU01005326 JAT02381.1 NEVH01016331 PNF25532.1 UFQT01000234 SSX22148.1 CH477634 EAT38026.1 PYGN01000107 PSN54116.1 GECZ01017810 JAS51959.1 GBGD01002788 JAC86101.1 JXUM01140342 JXUM01140343 JXUM01140344 JXUM01140345 KQ569047 KXJ68784.1 GBHO01037354 GBRD01013668 GDHC01012314 JAG06250.1 JAG52158.1 JAQ06315.1 AJWK01012785 ACPB03001521 GAMC01021703 JAB84852.1 CM000364 EDX14679.1 CVRI01000008 CRK88534.1 GBXI01006914 JAD07378.1 GEDC01022360 GEDC01021910 JAS14938.1 JAS15388.1 GDHF01023585 GDHF01013384 JAI28729.1 JAI38930.1 GAKP01020027 GAKP01020024 JAC38928.1 AXCN02000023 JRES01001623 KNC21364.1 AXCM01003710 AXCM01003711 CCAG010023115 CP012526 ALC45968.1 APCN01002026 AAAB01008987 EAL38486.3 CH940652 EDW59094.2 CH480837 EDW49069.1 AE014297 AY061370 AAF56738.2 AAL28918.1 KK855175 PTY21286.1 CH902617 EDV43872.1 CH933806 EDW16814.2 CH916374 EDV91032.1 CH964232 EDW80883.1 CH479186 EDW38853.1 CH954182 EDV53384.1 CM000070 EAL27036.3 DS232275 EDS38921.1 OUUW01000005 SPP80715.1 DS235755 EEB16401.1 GGMS01014988 MBY84191.1 ABLF02031876 ABLF02031877 GGMR01007136 MBY19755.1 KRG02311.1 KRG02312.1 KQ435010 KZC13724.1 ADTU01026200 GL449017 EFN83505.1 KQ976445 KYM86099.1
Proteomes
UP000005204
UP000053240
UP000218220
UP000037510
UP000007151
UP000192223
+ More
UP000007266 UP000019118 UP000030742 UP000235965 UP000008820 UP000245037 UP000078200 UP000069940 UP000249989 UP000092461 UP000015103 UP000092445 UP000000304 UP000183832 UP000075886 UP000037069 UP000075900 UP000075881 UP000075885 UP000076408 UP000075883 UP000075920 UP000092444 UP000092553 UP000076407 UP000075840 UP000075884 UP000095301 UP000075882 UP000007062 UP000075902 UP000008792 UP000001292 UP000075903 UP000000803 UP000075880 UP000095300 UP000075901 UP000007801 UP000069272 UP000009192 UP000001070 UP000007798 UP000192221 UP000008744 UP000008711 UP000001819 UP000092443 UP000002320 UP000268350 UP000009046 UP000007819 UP000076502 UP000005205 UP000008237 UP000078540
UP000007266 UP000019118 UP000030742 UP000235965 UP000008820 UP000245037 UP000078200 UP000069940 UP000249989 UP000092461 UP000015103 UP000092445 UP000000304 UP000183832 UP000075886 UP000037069 UP000075900 UP000075881 UP000075885 UP000076408 UP000075883 UP000075920 UP000092444 UP000092553 UP000076407 UP000075840 UP000075884 UP000095301 UP000075882 UP000007062 UP000075902 UP000008792 UP000001292 UP000075903 UP000000803 UP000075880 UP000095300 UP000075901 UP000007801 UP000069272 UP000009192 UP000001070 UP000007798 UP000192221 UP000008744 UP000008711 UP000001819 UP000092443 UP000002320 UP000268350 UP000009046 UP000007819 UP000076502 UP000005205 UP000008237 UP000078540
Pfam
PF16027 DUF4786
Interpro
IPR031983
DUF4786
ProteinModelPortal
H9JC79
A0A0N1IGS6
I4DK90
A0A2H1VA69
A0A2A4K5B7
A0A0L7KMD6
+ More
A0A212F855 A0A1Y1KKL0 A0A1W4XA86 D6WVH8 N6TPB6 A0A1B6JT32 A0A2J7QAD6 A0A336M7X7 Q16U16 A0A2P8ZC83 A0A1B6FP74 A0A069DQW7 A0A1A9VJ25 A0A182G2Y5 A0A0A9WN19 A0A1B0CHW5 T1HLP8 A0A1A9Z7B4 W8AGT2 B4QY31 A0A1J1HPY7 A0A0A1X9P2 A0A1B6CNH4 A0A0K8VK24 A0A034VBJ7 A0A182QGK7 A0A0L0BQ79 A0A182R666 A0A182JWY5 A0A182P571 A0A182YAP4 A0A182MDE4 A0A182VZ74 A0A1B0FP45 A0A0M5J4N2 A0A182X0A8 A0A182HZZ8 A0A182N431 A0A1I8NB81 A0A182LR29 Q380S4 A0A182U967 B4M618 B4IGV1 A0A182VLE4 Q9VB07 A0A182JF85 A0A1I8Q6C0 A0A1I8MK34 A0A2R7WNT5 A0A182SLY3 B3LX33 A0A182F968 B4KAS7 B4JU62 B4NAR9 A0A1W4V8X7 B4GNX0 B3P5Y4 Q29BG2 A0A1A9Y2J8 B0X1Z1 A0A3B0JF15 E0VSP5 A0A2S2R2C5 J9KA21 A0A2S2NRH9 A0A0Q9XDR7 A0A154PQV0 A0A158NU45 E2BLF3 A0A151I557
A0A212F855 A0A1Y1KKL0 A0A1W4XA86 D6WVH8 N6TPB6 A0A1B6JT32 A0A2J7QAD6 A0A336M7X7 Q16U16 A0A2P8ZC83 A0A1B6FP74 A0A069DQW7 A0A1A9VJ25 A0A182G2Y5 A0A0A9WN19 A0A1B0CHW5 T1HLP8 A0A1A9Z7B4 W8AGT2 B4QY31 A0A1J1HPY7 A0A0A1X9P2 A0A1B6CNH4 A0A0K8VK24 A0A034VBJ7 A0A182QGK7 A0A0L0BQ79 A0A182R666 A0A182JWY5 A0A182P571 A0A182YAP4 A0A182MDE4 A0A182VZ74 A0A1B0FP45 A0A0M5J4N2 A0A182X0A8 A0A182HZZ8 A0A182N431 A0A1I8NB81 A0A182LR29 Q380S4 A0A182U967 B4M618 B4IGV1 A0A182VLE4 Q9VB07 A0A182JF85 A0A1I8Q6C0 A0A1I8MK34 A0A2R7WNT5 A0A182SLY3 B3LX33 A0A182F968 B4KAS7 B4JU62 B4NAR9 A0A1W4V8X7 B4GNX0 B3P5Y4 Q29BG2 A0A1A9Y2J8 B0X1Z1 A0A3B0JF15 E0VSP5 A0A2S2R2C5 J9KA21 A0A2S2NRH9 A0A0Q9XDR7 A0A154PQV0 A0A158NU45 E2BLF3 A0A151I557
Ontologies
Topology
SignalP
Position: 1 - 21,
Likelihood: 0.986875
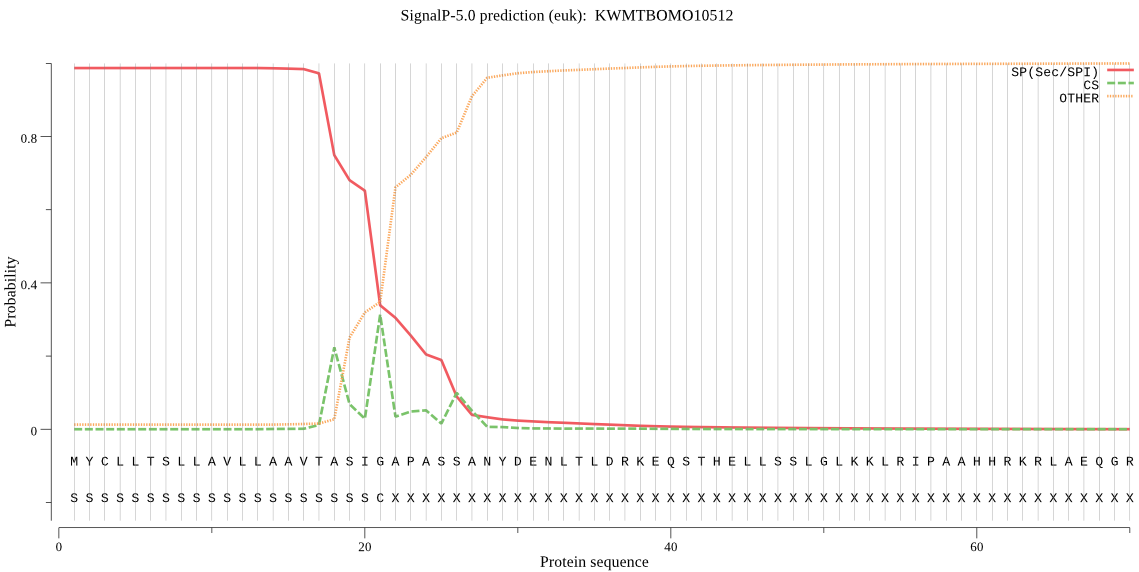
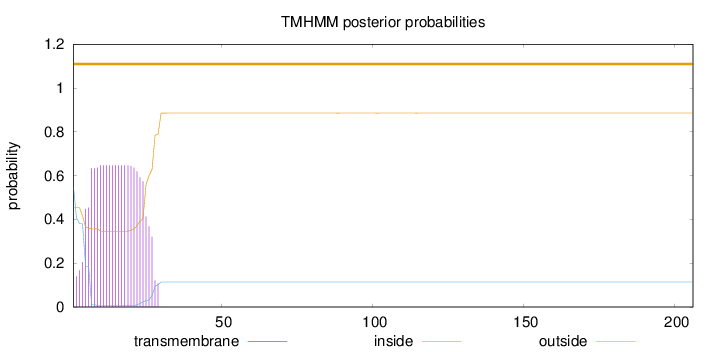
Length:
206
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
14.18717
Exp number, first 60 AAs:
14.18599
Total prob of N-in:
0.54683
POSSIBLE N-term signal
sequence
outside
1 - 206
Population Genetic Test Statistics
Pi
290.382265
Theta
160.887039
Tajima's D
2.059606
CLR
0.169279
CSRT
0.892105394730264
Interpretation
Uncertain
