Gene
KWMTBOMO10509 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006986
Annotation
ribosomal_protein_L22_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.632
Sequence
CDS
ATGGCAGTAACAAAGAAACCGGTGGCTAAGAAAGCCCAATTGCACCAGAAGACTGGCAAAAAAGGAGTGAAGGGTGGCAAAATCCGCGGCAAAGGCATCAAGCGGAAGATTAACTTAAAATTTACAATCGACTGCACGCATCCCGCTGAGGATAGTATCTTAGACGTAGGTAACTTCGAGAAATACTTGAAGGAACACGTCAAAGTTGAGGGCAAAACAAATAACCTAAGCAATCACGTTGTCGTCGCCAGGGATAAGACGAAAGTCGCTATCACCGCAGACATTCCTTTTTCAAAGAGGTACCTGAAGTATTTAACAAAACGTTACCTCAAGAAGAACAATTTGCGTGACTGGCTTCGAGTGGTGGCCTCCGCTCATGACGCTTACGAACTGCGCTACTTCAACATCAACGCTGACAGCGATAACGAAGACAATGAGGATTAA
Protein
MAVTKKPVAKKAQLHQKTGKKGVKGGKIRGKGIKRKINLKFTIDCTHPAEDSILDVGNFEKYLKEHVKVEGKTNNLSNHVVVARDKTKVAITADIPFSKRYLKYLTKRYLKKNNLRDWLRVVASAHDAYELRYFNINADSDNEDNED
Summary
Uniprot
Pubmed
EMBL
BABH01015035
AY769291
AAV34833.1
GDQN01005820
GDQN01005077
JAT85234.1
+ More
JAT85977.1 NWSH01000822 PCG74037.1 GBRD01001535 JAG64286.1 AF400188 ODYU01002382 AAK92160.1 SOQ39809.1 RBVL01000305 RKO09523.1 AK402425 BAM19047.1 KQ460711 KPJ12671.1 AK401144 KQ459603 BAM17766.1 KPI93107.1 HQ424767 ADT80711.1 GU084268 ACY95301.1 JF265060 AEL28891.1 JTDY01000561 KOB76676.1 MG992466 AXY94904.1 GAIX01009027 JAA83533.1 AGBW02010224 OWR48891.1 RSAL01000054 RVE50011.1 NIVC01000633 PAA79445.1 NIVC01004126 NIVC01000940 PAA48512.1 PAA74540.1 NIVC01004210 NIVC01004063 NIVC01002600 NIVC01001122 PAA48193.1 PAA48764.1 PAA56611.1 PAA71990.1 KC989881 KT754417 AGN29654.1 ALS04251.1 DS235076 EEB11426.1 GFTR01004420 JAW12006.1 GBGD01002505 JAC86384.1 GEMB01000159 JAS02954.1 KT754416 ALS04250.1 KT755237 ALS05071.1 ACPB03016514 GAHY01001353 JAA76157.1 JP593131 AER92516.1 RAZU01000078 RLQ75028.1
JAT85977.1 NWSH01000822 PCG74037.1 GBRD01001535 JAG64286.1 AF400188 ODYU01002382 AAK92160.1 SOQ39809.1 RBVL01000305 RKO09523.1 AK402425 BAM19047.1 KQ460711 KPJ12671.1 AK401144 KQ459603 BAM17766.1 KPI93107.1 HQ424767 ADT80711.1 GU084268 ACY95301.1 JF265060 AEL28891.1 JTDY01000561 KOB76676.1 MG992466 AXY94904.1 GAIX01009027 JAA83533.1 AGBW02010224 OWR48891.1 RSAL01000054 RVE50011.1 NIVC01000633 PAA79445.1 NIVC01004126 NIVC01000940 PAA48512.1 PAA74540.1 NIVC01004210 NIVC01004063 NIVC01002600 NIVC01001122 PAA48193.1 PAA48764.1 PAA56611.1 PAA71990.1 KC989881 KT754417 AGN29654.1 ALS04251.1 DS235076 EEB11426.1 GFTR01004420 JAW12006.1 GBGD01002505 JAC86384.1 GEMB01000159 JAS02954.1 KT754416 ALS04250.1 KT755237 ALS05071.1 ACPB03016514 GAHY01001353 JAA76157.1 JP593131 AER92516.1 RAZU01000078 RLQ75028.1
Proteomes
PRIDE
Gene 3D
ProteinModelPortal
PDB
3J92
E-value=1.73746e-22,
Score=256
Ontologies
GO
PANTHER
Topology
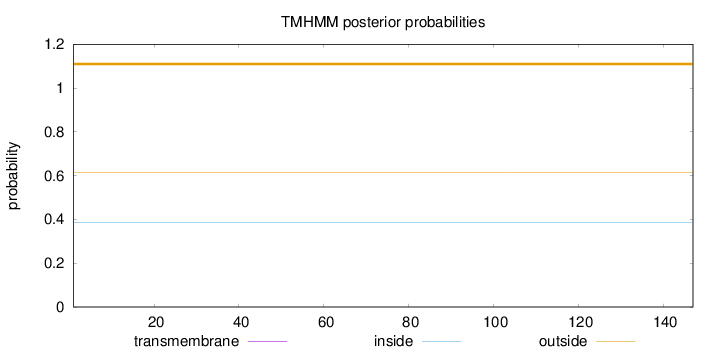
Length:
147
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00134
Exp number, first 60 AAs:
0
Total prob of N-in:
0.38645
outside
1 - 147
Population Genetic Test Statistics
Pi
207.078587
Theta
186.520957
Tajima's D
0.292632
CLR
0
CSRT
0.450277486125694
Interpretation
Uncertain
