Gene
KWMTBOMO10508 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007121
Annotation
PREDICTED:_probable_aconitate_hydratase?_mitochondrial_[Papilio_xuthus]
Full name
Aconitate hydratase, mitochondrial
Location in the cell
Mitochondrial Reliability : 3.767
Sequence
CDS
ATGGCGCACTGTATGAGAGTTTTGCATGGCCAGGGTAGCAGGACAAGGGTGGTACTCTCTGAAATCCAACAAAGATGTTTCAGCGTATCTCCATTGACCGCCGCCGCGGCCCAGGTGGCGATGTCCAAGTTCGACAAGGTACCCTTGCCTTACGAAAAGTTGACGAAGAATTTAGAGGTGGTTAAGAAGAGGCTAGGGAGAGAATTGACCTTGTCTGAGAAAATCCTATATTCTCACTTGGATGACCCCAAAGGACAGGAAATTGAACGCGGCGCAAGTTATCTCCGCCTGCGTCCCGACCGTGTGGCCATGCAAGACGCCACTGCACAAATGGCAATGTTACAATTTATCTCTTCCGGCCTTCCCCGTGTAGCTGTACCGTCCACCATTCATTGTGATCACTTGATCGAAGCCCAAGTCGGAGGCGAGAAGGATTTGGCCAGGGCTAAGGACCTCAACAAAGAAGTATACAAGTTTTTAGAGACTGCCGGAGCTAAATATGGAGTCGGTTTCTGGAAGCCCGGCTCTGGTATTATCCATCAGATCATTCTGGAGAACTATGCGTTCCCTGGTCTACTCATGATCGGCACAGACTCCCACACACCTAATGGAGGTGGCCTTGGTGGATTGTGTATTGGTGTCGGTGGTGCTGATGCTGTGGACGTAATGGCCAACATCCCGTGGGAGCTGAAGTGTCCTAAAGTTATCGGCGTCAAATTGACCGGTAAACTCACCGGCTGGACGAGTCCGAAAGATGTGATCTTGAAGGTGGCCGGCATCCTGACTGTGAAGGGAGGTACCGGCGCCATCGTGGAGTACCACGGACCCGGGGTGGACCACATCTCGTGTACAGGCATGGCGACAATCTGCAACATGGGAGCTGAGATCGGTGCTACCACTAGCGTGTTCCCGTACAACTCCCGCATGGAAGCCTACCTTAAGTCGACCGGGCGCCACGACATCGCCGCCACCGCCAACTCCTACAAGCACCTGCTCACGCCCGACAACAAGGCACCCTATGACCAGTTGATCGAGATCGACCTGTCCACGCTGGAGCCCCACGTGAACGGGCCCTTCACTCCGGACCTCGCCAACCCTATCTCGAAGCTCGGCGAAGCGGCCAAGAAGAACGACTGGCCGGTGGACATCCGCGTCGGCCTCATCGGGTCCTGCACCAACTCCTCCTACGAAGACATGGGCAGGTGCGCCAGCATCGTCAAAGAGGCGATCAGTCACGGCCTGAAGTCTAAGATTCCGTTCAACGTGACCCCCGGGTCGGAGCAGGTCCGTGCCACCATCGAGAGGGACGGCATCGCCAAGACCCTCAGGGACTTCGGAGGCACCGTGCTGGCGAACGCGTGCGGCCCGTGCATCGGCCAGTGGGACCGCAAGGACGTGAAGAAGGGGGAGAAGAACACGATCGTGACGTCATACAACCGCAACTTCACCGGCAGGAACGACGCCAACCCCGCCACGCACTGCTTCGTCACCAGCCCCGAGCTCGTCACGGCGCTGTCCATCGCGGGCCGGCTTGACTTCAACCCGCTGAAGGACGAACTGACTGGCGCTGATGGAAAGAAATTCAAGTTGTCAGATCCGTTTGCCGACGAGCTCCCCTCCAAGGGCTTCGACCCCGGACAGGACACGTACGAGCACCCGCCCGCCGACGGATCCAAAGTGAAAGTTGACGTCTCACCGACATCCGACCGCCTTCAACTCCTCGAGCCGTTCGACAAGTGGGACGGCAAAGACCTCACCGACCTCACGATCCTGATCAAGGTGAAGGGGAAGTGCACCACCGACCACATCTCGGCGGCTGGACCCTGGCTGAAGTACAGAGGCCATCTCGACAACATCTCCAATAACATGTTTATTACGGCAACGAACGCAGAGAACGGCGAGCTGAACAAAGTCCGCAACCAGGTGACGGGCGAGTACGGGCCGGTGCCCGCCACGGCGCGCGCGTACAAGGCGGCCGGCGTGCGCTGGTGCGTCGTCGGCGACGAGAACTACGGCGAGGGCTCGTCCCGCGAGCACGCCGCGCTCGAGCCGCGACACCTCGGCGGCCGCGCCATCATCGTCAAGTCGTTCGCTAGAATCCACGAGACCAACTTGAAGAAACAAGGTATGCTGCCCCTCACGTTCGCCAACGCCGCTGACTACGACAAGATCAAGCCTGACGATAAAATCTCACTCCTTGGACTGAACAGCCTCGCTCCCGGCAAGCCAGTGGACTGCGAAATTAAGCACAAAGACGGCAGCACCGACCGCATCAAGCTCAACCATTCGTTGAACGACCAACAGATCAACTGGTTCAAGGCCGGCTCGGCGCTCAACAGAATGAAGGAAATCGCTTCCGGAAAGTGA
Protein
MAHCMRVLHGQGSRTRVVLSEIQQRCFSVSPLTAAAAQVAMSKFDKVPLPYEKLTKNLEVVKKRLGRELTLSEKILYSHLDDPKGQEIERGASYLRLRPDRVAMQDATAQMAMLQFISSGLPRVAVPSTIHCDHLIEAQVGGEKDLARAKDLNKEVYKFLETAGAKYGVGFWKPGSGIIHQIILENYAFPGLLMIGTDSHTPNGGGLGGLCIGVGGADAVDVMANIPWELKCPKVIGVKLTGKLTGWTSPKDVILKVAGILTVKGGTGAIVEYHGPGVDHISCTGMATICNMGAEIGATTSVFPYNSRMEAYLKSTGRHDIAATANSYKHLLTPDNKAPYDQLIEIDLSTLEPHVNGPFTPDLANPISKLGEAAKKNDWPVDIRVGLIGSCTNSSYEDMGRCASIVKEAISHGLKSKIPFNVTPGSEQVRATIERDGIAKTLRDFGGTVLANACGPCIGQWDRKDVKKGEKNTIVTSYNRNFTGRNDANPATHCFVTSPELVTALSIAGRLDFNPLKDELTGADGKKFKLSDPFADELPSKGFDPGQDTYEHPPADGSKVKVDVSPTSDRLQLLEPFDKWDGKDLTDLTILIKVKGKCTTDHISAAGPWLKYRGHLDNISNNMFITATNAENGELNKVRNQVTGEYGPVPATARAYKAAGVRWCVVGDENYGEGSSREHAALEPRHLGGRAIIVKSFARIHETNLKKQGMLPLTFANAADYDKIKPDDKISLLGLNSLAPGKPVDCEIKHKDGSTDRIKLNHSLNDQQINWFKAGSALNRMKEIASGK
Summary
Cofactor
[4Fe-4S] cluster
Similarity
Belongs to the aconitase/IPM isomerase family.
Feature
chain Aconitate hydratase, mitochondrial
Uniprot
A0A194PI65
A0A3S2TMC5
Q86GF8
A0A2A4JQI9
D6WWL1
A0A1Y1KNE0
+ More
A0A2J7RRZ9 J3JWT3 A0A067R825 A0A1W4X0G9 A0A0T6B6I7 K7IMY0 A0A232FK46 A0A0C9QFK7 A0A2A3EDI2 V9IHT5 A0A087ZXK3 A0A0J7NRU4 Q16KR4 A0A336K422 A0A023EW32 A0A1B6DJW1 A0A182GM09 A0A158NLP6 A0A151IU55 A0A3L8DIS0 Q17EL3 F4X2U7 A0A1B6H3E8 A0A182QJC6 A0A151WK30 A0A195BLF6 A0A182NLL1 A0A1J1I2U2 A0A026X5E9 A0A3Q0IZI1 A0A182H7F2 A0A182MK76 A0A182YPN0 A0A0M8ZWK8 E2A4Y0 A0A182W9X3 R4WKE5 A0A182JQG1 E2BSG2 A0A182IEM3 A0A182P0J0 A0A182UN50 A0A182L2B6 A0A182WTG5 A0A1S4GWF2 A0A182R397 T1DN98 A0A2M3ZZX0 A0A182TV65 A0A2H8TNH1 A0A1Q3G4U8 A0A0C5QDN7 A0A182F685 A0A2M3YXT4 A0A154P379 A0A2M4BEF5 A0A2M3YXT6 A0A2M4BEF8 A0A023EXU2 A0A2M4CW41 T1DHZ4 A0A084W3X9 Q7Q3F6 A0A182IXZ9 A0A2M4CUU9 A0A0A9X6E8 A0A023EXR0 U5EU23 T1PD24 A0A1I8N262 J9KAT0 A0A2M3YXV2 A0A0L7QQW8 A0A195FV98 A0A1L8EDF9 A0A151IAM2 A0A069DWY8 W5JMI4 A0A310S9H3 A0A0A1XE57 A0A1B0CX01 A0A1I8NML7 W8B5J2 A0A2P2HXE8 R4G408 A0A1L8DH08 A0A1L8DH00 A0A0P4VIL1 A0A0L0CD74 A0A034W2U8 Q29NT1 B4GJJ6 A0A023GLX2 E9GUX6
A0A2J7RRZ9 J3JWT3 A0A067R825 A0A1W4X0G9 A0A0T6B6I7 K7IMY0 A0A232FK46 A0A0C9QFK7 A0A2A3EDI2 V9IHT5 A0A087ZXK3 A0A0J7NRU4 Q16KR4 A0A336K422 A0A023EW32 A0A1B6DJW1 A0A182GM09 A0A158NLP6 A0A151IU55 A0A3L8DIS0 Q17EL3 F4X2U7 A0A1B6H3E8 A0A182QJC6 A0A151WK30 A0A195BLF6 A0A182NLL1 A0A1J1I2U2 A0A026X5E9 A0A3Q0IZI1 A0A182H7F2 A0A182MK76 A0A182YPN0 A0A0M8ZWK8 E2A4Y0 A0A182W9X3 R4WKE5 A0A182JQG1 E2BSG2 A0A182IEM3 A0A182P0J0 A0A182UN50 A0A182L2B6 A0A182WTG5 A0A1S4GWF2 A0A182R397 T1DN98 A0A2M3ZZX0 A0A182TV65 A0A2H8TNH1 A0A1Q3G4U8 A0A0C5QDN7 A0A182F685 A0A2M3YXT4 A0A154P379 A0A2M4BEF5 A0A2M3YXT6 A0A2M4BEF8 A0A023EXU2 A0A2M4CW41 T1DHZ4 A0A084W3X9 Q7Q3F6 A0A182IXZ9 A0A2M4CUU9 A0A0A9X6E8 A0A023EXR0 U5EU23 T1PD24 A0A1I8N262 J9KAT0 A0A2M3YXV2 A0A0L7QQW8 A0A195FV98 A0A1L8EDF9 A0A151IAM2 A0A069DWY8 W5JMI4 A0A310S9H3 A0A0A1XE57 A0A1B0CX01 A0A1I8NML7 W8B5J2 A0A2P2HXE8 R4G408 A0A1L8DH08 A0A1L8DH00 A0A0P4VIL1 A0A0L0CD74 A0A034W2U8 Q29NT1 B4GJJ6 A0A023GLX2 E9GUX6
EC Number
4.2.1.-
Pubmed
26354079
18362917
19820115
28004739
22516182
23537049
+ More
24845553 20075255 28648823 17510324 24945155 26483478 21347285 30249741 21719571 24508170 25244985 20798317 23691247 20966253 12364791 25702953 24330624 24438588 25401762 26823975 25315136 26334808 20920257 23761445 25830018 24495485 27129103 26108605 25348373 15632085 17994087 21292972
24845553 20075255 28648823 17510324 24945155 26483478 21347285 30249741 21719571 24508170 25244985 20798317 23691247 20966253 12364791 25702953 24330624 24438588 25401762 26823975 25315136 26334808 20920257 23761445 25830018 24495485 27129103 26108605 25348373 15632085 17994087 21292972
EMBL
KQ459603
KPI93106.1
RSAL01000054
RVE50012.1
AB105050
BAC65324.1
+ More
NWSH01000822 PCG74036.1 KQ971361 EFA08121.1 GEZM01083906 JAV60956.1 NEVH01000597 PNF43609.1 APGK01058660 BT127701 KB741291 KB631579 AEE62663.1 ENN70544.1 ERL84416.1 KK852872 KDR14559.1 LJIG01009490 KRT82976.1 NNAY01000130 OXU30697.1 GBYB01013263 JAG83030.1 KZ288288 PBC29352.1 JR048497 AEY60683.1 LBMM01002199 KMQ95155.1 CH477941 EAT34906.1 UFQS01000048 UFQT01000048 SSW98592.1 SSX18978.1 GAPW01000256 JAC13342.1 GEDC01011312 JAS25986.1 JXUM01073449 KQ562775 KXJ75145.1 ADTU01019627 KQ980968 KYN10979.1 QOIP01000007 RLU20340.1 CH477280 EAT44980.1 GL888591 EGI59293.1 GECZ01000560 JAS69209.1 AXCN02000614 KQ983020 KYQ48222.1 KQ976450 KYM85637.1 CVRI01000038 CRK93908.1 KK107019 EZA62649.1 JXUM01116117 JXUM01116118 JXUM01116119 JXUM01116120 AXCM01007919 KQ435834 KOX71536.1 GL436756 EFN71522.1 AK418050 BAN21265.1 GL450233 EFN81350.1 APCN01005209 AAAB01008964 GAMD01003080 JAA98510.1 GGFK01000793 MBW34114.1 GFXV01003377 MBW15182.1 GFDL01000213 JAV34832.1 KM609494 AJQ30118.1 GGFM01000328 MBW21079.1 KQ434796 KZC05590.1 GGFJ01002284 MBW51425.1 GGFM01000326 MBW21077.1 GGFJ01002283 MBW51424.1 GAPW01000254 JAC13344.1 GGFL01004880 MBW69058.1 GALA01001246 JAA93606.1 ATLV01020201 KE525295 KFB44923.1 EAA12184.4 GGFL01004881 MBW69059.1 GBHO01028388 GBRD01017760 GDHC01017684 JAG15216.1 JAG48067.1 JAQ00945.1 GAPW01000309 JAC13289.1 GANO01001669 JAB58202.1 KA646574 AFP61203.1 ABLF02022040 GGFM01000348 MBW21099.1 KQ414784 KOC61025.1 KQ981219 KYN44388.1 GFDG01002134 JAV16665.1 KQ978186 KYM96557.1 GBGD01000548 JAC88341.1 ADMH02000676 ETN65346.1 KQ768478 OAD53151.1 GBXI01005076 JAD09216.1 AJWK01033078 AJWK01033079 AJWK01033080 AJWK01033081 AJWK01033082 AJWK01033083 AJWK01033084 GAMC01014172 GAMC01014170 JAB92383.1 IACF01000700 LAB66458.1 ACPB03000177 GAHY01001755 JAA75755.1 GFDF01008351 JAV05733.1 GFDF01008352 JAV05732.1 GDKW01002818 JAI53777.1 JRES01000668 KNC29409.1 GAKP01010854 JAC48098.1 CH379058 EAL34563.2 CH479184 EDW36812.1 GBBM01000516 JAC34902.1 GL732567 EFX76569.1
NWSH01000822 PCG74036.1 KQ971361 EFA08121.1 GEZM01083906 JAV60956.1 NEVH01000597 PNF43609.1 APGK01058660 BT127701 KB741291 KB631579 AEE62663.1 ENN70544.1 ERL84416.1 KK852872 KDR14559.1 LJIG01009490 KRT82976.1 NNAY01000130 OXU30697.1 GBYB01013263 JAG83030.1 KZ288288 PBC29352.1 JR048497 AEY60683.1 LBMM01002199 KMQ95155.1 CH477941 EAT34906.1 UFQS01000048 UFQT01000048 SSW98592.1 SSX18978.1 GAPW01000256 JAC13342.1 GEDC01011312 JAS25986.1 JXUM01073449 KQ562775 KXJ75145.1 ADTU01019627 KQ980968 KYN10979.1 QOIP01000007 RLU20340.1 CH477280 EAT44980.1 GL888591 EGI59293.1 GECZ01000560 JAS69209.1 AXCN02000614 KQ983020 KYQ48222.1 KQ976450 KYM85637.1 CVRI01000038 CRK93908.1 KK107019 EZA62649.1 JXUM01116117 JXUM01116118 JXUM01116119 JXUM01116120 AXCM01007919 KQ435834 KOX71536.1 GL436756 EFN71522.1 AK418050 BAN21265.1 GL450233 EFN81350.1 APCN01005209 AAAB01008964 GAMD01003080 JAA98510.1 GGFK01000793 MBW34114.1 GFXV01003377 MBW15182.1 GFDL01000213 JAV34832.1 KM609494 AJQ30118.1 GGFM01000328 MBW21079.1 KQ434796 KZC05590.1 GGFJ01002284 MBW51425.1 GGFM01000326 MBW21077.1 GGFJ01002283 MBW51424.1 GAPW01000254 JAC13344.1 GGFL01004880 MBW69058.1 GALA01001246 JAA93606.1 ATLV01020201 KE525295 KFB44923.1 EAA12184.4 GGFL01004881 MBW69059.1 GBHO01028388 GBRD01017760 GDHC01017684 JAG15216.1 JAG48067.1 JAQ00945.1 GAPW01000309 JAC13289.1 GANO01001669 JAB58202.1 KA646574 AFP61203.1 ABLF02022040 GGFM01000348 MBW21099.1 KQ414784 KOC61025.1 KQ981219 KYN44388.1 GFDG01002134 JAV16665.1 KQ978186 KYM96557.1 GBGD01000548 JAC88341.1 ADMH02000676 ETN65346.1 KQ768478 OAD53151.1 GBXI01005076 JAD09216.1 AJWK01033078 AJWK01033079 AJWK01033080 AJWK01033081 AJWK01033082 AJWK01033083 AJWK01033084 GAMC01014172 GAMC01014170 JAB92383.1 IACF01000700 LAB66458.1 ACPB03000177 GAHY01001755 JAA75755.1 GFDF01008351 JAV05733.1 GFDF01008352 JAV05732.1 GDKW01002818 JAI53777.1 JRES01000668 KNC29409.1 GAKP01010854 JAC48098.1 CH379058 EAL34563.2 CH479184 EDW36812.1 GBBM01000516 JAC34902.1 GL732567 EFX76569.1
Proteomes
UP000053268
UP000283053
UP000218220
UP000007266
UP000235965
UP000019118
+ More
UP000030742 UP000027135 UP000192223 UP000002358 UP000215335 UP000242457 UP000005203 UP000036403 UP000008820 UP000069940 UP000249989 UP000005205 UP000078492 UP000279307 UP000007755 UP000075886 UP000075809 UP000078540 UP000075884 UP000183832 UP000053097 UP000079169 UP000075883 UP000076408 UP000053105 UP000000311 UP000075920 UP000075881 UP000008237 UP000075840 UP000075885 UP000075903 UP000075882 UP000076407 UP000075900 UP000075902 UP000069272 UP000076502 UP000030765 UP000007062 UP000075880 UP000095301 UP000007819 UP000053825 UP000078541 UP000078542 UP000000673 UP000092461 UP000095300 UP000015103 UP000037069 UP000001819 UP000008744 UP000000305
UP000030742 UP000027135 UP000192223 UP000002358 UP000215335 UP000242457 UP000005203 UP000036403 UP000008820 UP000069940 UP000249989 UP000005205 UP000078492 UP000279307 UP000007755 UP000075886 UP000075809 UP000078540 UP000075884 UP000183832 UP000053097 UP000079169 UP000075883 UP000076408 UP000053105 UP000000311 UP000075920 UP000075881 UP000008237 UP000075840 UP000075885 UP000075903 UP000075882 UP000076407 UP000075900 UP000075902 UP000069272 UP000076502 UP000030765 UP000007062 UP000075880 UP000095301 UP000007819 UP000053825 UP000078541 UP000078542 UP000000673 UP000092461 UP000095300 UP000015103 UP000037069 UP000001819 UP000008744 UP000000305
PRIDE
Interpro
IPR036008
Aconitase_4Fe-4S_dom
+ More
IPR018136 Aconitase_4Fe-4S_BS
IPR000573 AconitaseA/IPMdHydase_ssu_swvl
IPR015932 Aconitase_dom2
IPR006248 Aconitase_mito-like
IPR001030 Acoase/IPM_deHydtase_lsu_aba
IPR015928 Aconitase/3IPM_dehydase_swvl
IPR015931 Acnase/IPM_dHydase_lsu_aba_1/3
IPR007529 Znf_HIT
IPR018136 Aconitase_4Fe-4S_BS
IPR000573 AconitaseA/IPMdHydase_ssu_swvl
IPR015932 Aconitase_dom2
IPR006248 Aconitase_mito-like
IPR001030 Acoase/IPM_deHydtase_lsu_aba
IPR015928 Aconitase/3IPM_dehydase_swvl
IPR015931 Acnase/IPM_dHydase_lsu_aba_1/3
IPR007529 Znf_HIT
SUPFAM
SSF53732
SSF53732
Gene 3D
ProteinModelPortal
A0A194PI65
A0A3S2TMC5
Q86GF8
A0A2A4JQI9
D6WWL1
A0A1Y1KNE0
+ More
A0A2J7RRZ9 J3JWT3 A0A067R825 A0A1W4X0G9 A0A0T6B6I7 K7IMY0 A0A232FK46 A0A0C9QFK7 A0A2A3EDI2 V9IHT5 A0A087ZXK3 A0A0J7NRU4 Q16KR4 A0A336K422 A0A023EW32 A0A1B6DJW1 A0A182GM09 A0A158NLP6 A0A151IU55 A0A3L8DIS0 Q17EL3 F4X2U7 A0A1B6H3E8 A0A182QJC6 A0A151WK30 A0A195BLF6 A0A182NLL1 A0A1J1I2U2 A0A026X5E9 A0A3Q0IZI1 A0A182H7F2 A0A182MK76 A0A182YPN0 A0A0M8ZWK8 E2A4Y0 A0A182W9X3 R4WKE5 A0A182JQG1 E2BSG2 A0A182IEM3 A0A182P0J0 A0A182UN50 A0A182L2B6 A0A182WTG5 A0A1S4GWF2 A0A182R397 T1DN98 A0A2M3ZZX0 A0A182TV65 A0A2H8TNH1 A0A1Q3G4U8 A0A0C5QDN7 A0A182F685 A0A2M3YXT4 A0A154P379 A0A2M4BEF5 A0A2M3YXT6 A0A2M4BEF8 A0A023EXU2 A0A2M4CW41 T1DHZ4 A0A084W3X9 Q7Q3F6 A0A182IXZ9 A0A2M4CUU9 A0A0A9X6E8 A0A023EXR0 U5EU23 T1PD24 A0A1I8N262 J9KAT0 A0A2M3YXV2 A0A0L7QQW8 A0A195FV98 A0A1L8EDF9 A0A151IAM2 A0A069DWY8 W5JMI4 A0A310S9H3 A0A0A1XE57 A0A1B0CX01 A0A1I8NML7 W8B5J2 A0A2P2HXE8 R4G408 A0A1L8DH08 A0A1L8DH00 A0A0P4VIL1 A0A0L0CD74 A0A034W2U8 Q29NT1 B4GJJ6 A0A023GLX2 E9GUX6
A0A2J7RRZ9 J3JWT3 A0A067R825 A0A1W4X0G9 A0A0T6B6I7 K7IMY0 A0A232FK46 A0A0C9QFK7 A0A2A3EDI2 V9IHT5 A0A087ZXK3 A0A0J7NRU4 Q16KR4 A0A336K422 A0A023EW32 A0A1B6DJW1 A0A182GM09 A0A158NLP6 A0A151IU55 A0A3L8DIS0 Q17EL3 F4X2U7 A0A1B6H3E8 A0A182QJC6 A0A151WK30 A0A195BLF6 A0A182NLL1 A0A1J1I2U2 A0A026X5E9 A0A3Q0IZI1 A0A182H7F2 A0A182MK76 A0A182YPN0 A0A0M8ZWK8 E2A4Y0 A0A182W9X3 R4WKE5 A0A182JQG1 E2BSG2 A0A182IEM3 A0A182P0J0 A0A182UN50 A0A182L2B6 A0A182WTG5 A0A1S4GWF2 A0A182R397 T1DN98 A0A2M3ZZX0 A0A182TV65 A0A2H8TNH1 A0A1Q3G4U8 A0A0C5QDN7 A0A182F685 A0A2M3YXT4 A0A154P379 A0A2M4BEF5 A0A2M3YXT6 A0A2M4BEF8 A0A023EXU2 A0A2M4CW41 T1DHZ4 A0A084W3X9 Q7Q3F6 A0A182IXZ9 A0A2M4CUU9 A0A0A9X6E8 A0A023EXR0 U5EU23 T1PD24 A0A1I8N262 J9KAT0 A0A2M3YXV2 A0A0L7QQW8 A0A195FV98 A0A1L8EDF9 A0A151IAM2 A0A069DWY8 W5JMI4 A0A310S9H3 A0A0A1XE57 A0A1B0CX01 A0A1I8NML7 W8B5J2 A0A2P2HXE8 R4G408 A0A1L8DH08 A0A1L8DH00 A0A0P4VIL1 A0A0L0CD74 A0A034W2U8 Q29NT1 B4GJJ6 A0A023GLX2 E9GUX6
PDB
1AMJ
E-value=0,
Score=3083
Ontologies
PATHWAY
00020
Citrate cycle (TCA cycle) - Bombyx mori (domestic silkworm)
00630 Glyoxylate and dicarboxylate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01210 2-Oxocarboxylic acid metabolism - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
00630 Glyoxylate and dicarboxylate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01210 2-Oxocarboxylic acid metabolism - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
GO
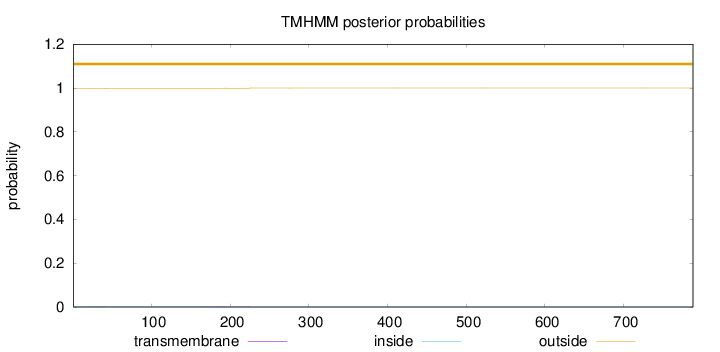
Topology
Subcellular location
Mitochondrion
Length:
788
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03267
Exp number, first 60 AAs:
0.00707
Total prob of N-in:
0.00163
outside
1 - 788
Population Genetic Test Statistics
Pi
235.168141
Theta
162.808271
Tajima's D
1.359197
CLR
0.349329
CSRT
0.752662366881656
Interpretation
Uncertain
