Pre Gene Modal
BGIBMGA006987
Annotation
PREDICTED:_exocyst_complex_component_6_[Papilio_polytes]
Full name
Exocyst complex component
+ More
Exocyst complex component 6
Exocyst complex component 6
Alternative Name
Exocyst complex component Sec15
Location in the cell
Cytoplasmic Reliability : 3.059
Sequence
CDS
ATGAATGCAACGATTCAGGAAATCGAAGGTATAGATGATTATTGGGGGCCGGCATTCCGATCCGTTTACGAAGGTGAAGGGCACGAAGCATTCGTGCAACAATTAGATGATCGAATAAAGCAGCACGATAAGGAAATTGAGAAATTGTGCAATTTTCACTATCAGGGTTTCATAGATTCTATAAGAGAATTGCTGCAAGTCCGTTCTCATGCTGAAGAATTGCATGCTGAAATATCTAATGTAGATGCTAATGTCAAAGAGACTACTGAAGCACTGTGCATTAGAGCAGACGAATTGATCAGGGCTCGAAGAGTCGAACTGAACATTGCAGCTACCATTGAAAAAATGGAACTGTGTTTACCTCTCCTCACTACTTACTCCAAACTCAAATCCCAAGTAGAAGCGAAGAGATATTATCCAGCACTTAAAACTCTAGAGCAACTCGAACATGTGCTACTGCCGCGAGTCGGTCCATACAAATGGTGCGCACAGATATCTTCCGATATACCACGATTGAGGCAGGCCATACGAGATGCTTCAATGGCCGATTTGAGGGACTTTCTAGAGAACATTCGTAGGCTTAGCCCTCAGGTCGGTGCGATGGCTCTGAAGCAAACTCAAGATATGATGGGCAGAAGTTTAGCTAACATAATCAGTAAAAAAGATAAATCACAGGGCTTGACGGCTTCGGACGGTCCTCAGTGTCCCCACGAGCTGGTAGACTTCAGTCCTCTGCATAGATGTCTGCACATCCACACCGTGCTAGGAGCCAAGAACGACTTCATACAGTATTACAGATCTCAGCGCAGGCAACAAGCCCGCCTCGTATTGATTCCGTCGTCTGGCAATCTCCACGATTCAATCCAGGGCATCAGGAACTATCTGCACGCAGTCCTCGGCTTCTTCATACTCGAGGAGCACTTGCTGACCGCCGGGGCCGGGCTGGTGTCCAAGGACTGGTTGCTGGACACCTGGGGCATGTCGGTGACCAAGGTGGCGTCGACACTCCGGACCAACACGTCGCTAGTAACCGACCCTAACTTGATGCTTAAAATCAAGGATCAAATTATGCTGTTCATAAACACTCTTAAATGCTACGGGCTGCCGACGGACCCGCTGCCGCTGCTGCTGCAGGAGATGGCCGAGCACTACACCGAGGTGCTGATGCAGCGGTGGGTCGGCGTGTTCCGGGACATACTCGACAACGGATCCTTCCTGCCCATCGAGGTTCAAAATCAAGAGGAATATGACAGCGTGATGGACAAGTTCCCTTACGACGGCGAAGAGCTGGAGACGCAACCGTTCCCCAGGAAGTTTCCGTTCTCGTCCCTGGTGACGTCCGTGTACGTGCAAGTGAAGGAGTTCATATACGCGTGGCTGATGTACTCGGCGGGGCTGGGGCTGGGCGGCGAGCGGCGGGCGGGCGCGGCGCGGCACTCCGCCTCGCTGCTGCTCAGCCGCTCCTTCGCCGGCTGTCTGTCGGCGCTGTTCCGCCGCCCGCTGCCGCTCGTGCAGCTCGTGCAGATAATAGTGGATACGCAGTATTTAGAAGACGCCACGGTCTGTCTGTACGAATTCATCAGCAACATAACGGGCTCGGAACTCGTGACCACACAGGCCGCGGGCAGCATGTTCCAGGCAGCTCGCGACGACGCCGAGCAGCAGATATGCGACAAGCTGGAGAAGAAGGTGGACGAATTCCTCGACTTAGAAAACTATGACTGGCTCTTAGTGGAGCCCATCGGACAAGCCTCGTCCTTCGTGACGGACATGCTCAGCTACTTATCGGGGGTGCTGACGTCACTGGAGCAGCTGCCCGAGCGAGCCAGGGTCGCGGCGGTTCGGGCGGCAACGAACCGCATCGCCACGCGACTCCGCGCCCTGCTGCTGGACCCGGCCGTCAAGCAGATCTCGTCCGGCGCGCTGTACCAACTCGACCTGGACGTGATCCAGTGCGAGCAGTTCGCGGCCGGCGAGCCTGTCCCCGGCCTCCGGGAGGGGGAACTGCTCGAGCACTTCGCTAGTTTGAGGCAACTACTGGATCTGATCACGGGCTGGGACTGGTCGTCGTACCTCCACGACGTCGGCATCGACGGCGGAAAATACACGCTCGTTACGCCCAAAGACGCGGCCACCCTGCTCGAGAAACTAAAGGAGGCCGAGCAGAAGTCTTCCGTCTTCTCTGTTCTGAAGAAAAACGAGAGAGACAGACGTAAGCTCCTCGACACCGTCCTGAAGCAACTGAAGCAGTTGCAGAATCAGGATTCTTGA
Protein
MNATIQEIEGIDDYWGPAFRSVYEGEGHEAFVQQLDDRIKQHDKEIEKLCNFHYQGFIDSIRELLQVRSHAEELHAEISNVDANVKETTEALCIRADELIRARRVELNIAATIEKMELCLPLLTTYSKLKSQVEAKRYYPALKTLEQLEHVLLPRVGPYKWCAQISSDIPRLRQAIRDASMADLRDFLENIRRLSPQVGAMALKQTQDMMGRSLANIISKKDKSQGLTASDGPQCPHELVDFSPLHRCLHIHTVLGAKNDFIQYYRSQRRQQARLVLIPSSGNLHDSIQGIRNYLHAVLGFFILEEHLLTAGAGLVSKDWLLDTWGMSVTKVASTLRTNTSLVTDPNLMLKIKDQIMLFINTLKCYGLPTDPLPLLLQEMAEHYTEVLMQRWVGVFRDILDNGSFLPIEVQNQEEYDSVMDKFPYDGEELETQPFPRKFPFSSLVTSVYVQVKEFIYAWLMYSAGLGLGGERRAGAARHSASLLLSRSFAGCLSALFRRPLPLVQLVQIIVDTQYLEDATVCLYEFISNITGSELVTTQAAGSMFQAARDDAEQQICDKLEKKVDEFLDLENYDWLLVEPIGQASSFVTDMLSYLSGVLTSLEQLPERARVAAVRAATNRIATRLRALLLDPAVKQISSGALYQLDLDVIQCEQFAAGEPVPGLREGELLEHFASLRQLLDLITGWDWSSYLHDVGIDGGKYTLVTPKDAATLLEKLKEAEQKSSVFSVLKKNERDRRKLLDTVLKQLKQLQNQDS
Summary
Description
Component of the exocyst complex involved in the docking of exocytic vesicles with fusion sites on the plasma membrane.
Subunit
The exocyst complex is composed of Sec3/Exoc1, Sec5/Exoc2, Sec6/Exoc3, Sec8/Exoc4, Sec10/Exoc5, Sec15/Exoc6, Exo70/Exoc7 and Exo84/Exoc8 (By similarity). Interacts with RAB3, RAB8, RAB11 and RAB27.
Similarity
Belongs to the SEC15 family.
Belongs to the peptidase C14A family.
Belongs to the peptidase C14A family.
Keywords
3D-structure
Coiled coil
Complete proteome
Exocytosis
Protein transport
Reference proteome
Transport
Feature
chain Exocyst complex component 6
Uniprot
H9JBU2
A0A2A4JPP3
A0A2H1W243
A0A3S2M3B5
A0A194PPP6
A0A212EUV6
+ More
A0A0N1PGR5 A0A2J7Q098 T1ICC1 A0A1B6DQE0 A0A1B6KDJ2 A0A0V0G758 A0A023F4S5 A0A0C9QWG4 A0A0A9XMQ2 E2BQM2 A0A1B6FRM1 A0A067QT00 K7IYQ6 A0A1B6E1B3 A0A154PCZ2 A0A088ADK2 A0A1B6C9M8 A0A1B6MIK1 A0A224XIN4 A0A0J7KAQ1 A0A026WDL2 A0A2A3EPV3 E2AW91 A0A0A9XNN5 A0A0L7RGP6 A0A3L8DWW5 A0A0N0BC08 A0A1W4X769 A0A232FFX4 F4WR05 A0A1Y1MMF6 A0A151IVS7 A0A158NXX0 A0A151HZ51 A0A195FWC6 A0A151WIG8 D6X4X0 E0VNA5 A0A1L8DVT5 B0WEG6 U4U091 A0A1Q3FFH0 Q17EP0 A0A084WMD1 A0A1S4F5K5 A0A182J5G8 A0A182Y2G0 A0A1I8MEH2 T1PJV5 A0A182G4S8 A0A182X4M8 A0A182M0K3 A0A182RDK0 A0A182NI62 A0A182K1S6 A0A182WBM2 W5J9E9 A0A182V8N7 A0A182TGC6 A0A182I4M6 A0A182FDB4 Q7Q546 A0A2S2QIL0 A0A182PDJ7 A0A182QQG8 A0A1I8PYW4 A0A1B0G9K3 A0A1A9ZUK1 A0A1A9UZY9 A0A1B0C8E7 A0A1A9XSW6 A0A1B0BKM5 J9K5H0 A0A2S2PUZ7 A0A1A9W9E0 B4JFJ0 A0A0P5R9Z7 A0A0N8ENA1 A0A0P5USB6 A0A336KYV7 Q296V4 B4GEL1 A0A0P5AH24 A0A0P4ZLA6 W8B9F1 A0A0L0CJZ9 A0A0P5V262 E9G3C4 A0A0K8UC27 B4R147 Q9VDE6 A0A3B0JMC0 A0A034W611 B3M2W2
A0A0N1PGR5 A0A2J7Q098 T1ICC1 A0A1B6DQE0 A0A1B6KDJ2 A0A0V0G758 A0A023F4S5 A0A0C9QWG4 A0A0A9XMQ2 E2BQM2 A0A1B6FRM1 A0A067QT00 K7IYQ6 A0A1B6E1B3 A0A154PCZ2 A0A088ADK2 A0A1B6C9M8 A0A1B6MIK1 A0A224XIN4 A0A0J7KAQ1 A0A026WDL2 A0A2A3EPV3 E2AW91 A0A0A9XNN5 A0A0L7RGP6 A0A3L8DWW5 A0A0N0BC08 A0A1W4X769 A0A232FFX4 F4WR05 A0A1Y1MMF6 A0A151IVS7 A0A158NXX0 A0A151HZ51 A0A195FWC6 A0A151WIG8 D6X4X0 E0VNA5 A0A1L8DVT5 B0WEG6 U4U091 A0A1Q3FFH0 Q17EP0 A0A084WMD1 A0A1S4F5K5 A0A182J5G8 A0A182Y2G0 A0A1I8MEH2 T1PJV5 A0A182G4S8 A0A182X4M8 A0A182M0K3 A0A182RDK0 A0A182NI62 A0A182K1S6 A0A182WBM2 W5J9E9 A0A182V8N7 A0A182TGC6 A0A182I4M6 A0A182FDB4 Q7Q546 A0A2S2QIL0 A0A182PDJ7 A0A182QQG8 A0A1I8PYW4 A0A1B0G9K3 A0A1A9ZUK1 A0A1A9UZY9 A0A1B0C8E7 A0A1A9XSW6 A0A1B0BKM5 J9K5H0 A0A2S2PUZ7 A0A1A9W9E0 B4JFJ0 A0A0P5R9Z7 A0A0N8ENA1 A0A0P5USB6 A0A336KYV7 Q296V4 B4GEL1 A0A0P5AH24 A0A0P4ZLA6 W8B9F1 A0A0L0CJZ9 A0A0P5V262 E9G3C4 A0A0K8UC27 B4R147 Q9VDE6 A0A3B0JMC0 A0A034W611 B3M2W2
Pubmed
19121390
26354079
22118469
25474469
25401762
20798317
+ More
24845553 20075255 24508170 30249741 28648823 21719571 28004739 21347285 18362917 19820115 20566863 23537049 17510324 24438588 25244985 25315136 26483478 20920257 23761445 12364791 14747013 17210077 17994087 15632085 24495485 26108605 21292972 10731132 12537572 16155582 25348373
24845553 20075255 24508170 30249741 28648823 21719571 28004739 21347285 18362917 19820115 20566863 23537049 17510324 24438588 25244985 25315136 26483478 20920257 23761445 12364791 14747013 17210077 17994087 15632085 24495485 26108605 21292972 10731132 12537572 16155582 25348373
EMBL
BABH01015034
BABH01015035
NWSH01000822
PCG74035.1
ODYU01005861
SOQ47160.1
+ More
RSAL01000054 RVE50013.1 KQ459603 KPI93105.1 AGBW02012291 OWR45272.1 KQ460711 KPJ12669.1 NEVH01019998 PNF21996.1 ACPB03019765 GEDC01009475 JAS27823.1 GEBQ01030470 JAT09507.1 GECL01002286 JAP03838.1 GBBI01002709 JAC16003.1 GBYB01000060 JAG69827.1 GBHO01039771 GBHO01022367 GBHO01022365 JAG03833.1 JAG21237.1 JAG21239.1 GL449784 EFN81987.1 GECZ01016907 JAS52862.1 KK853313 KDR08619.1 GEDC01005583 JAS31715.1 KQ434875 KZC09765.1 GEDC01027115 JAS10183.1 GEBQ01004190 JAT35787.1 GFTR01007984 JAW08442.1 LBMM01010688 KMQ87316.1 KK107295 EZA53099.1 KZ288206 PBC33071.1 GL443262 EFN62313.1 GBHO01022363 GBHO01022361 GBHO01022360 JAG21241.1 JAG21243.1 JAG21244.1 KQ414596 KOC70025.1 QOIP01000003 RLU24964.1 KQ435944 KOX68212.1 NNAY01000292 OXU29420.1 GL888280 EGI63363.1 GEZM01031081 JAV85076.1 KQ980882 KYN11946.1 ADTU01003649 ADTU01003650 KQ976709 KYM77042.1 KQ981261 KYN44139.1 KQ983089 KYQ47616.1 KQ971381 EEZ97223.1 DS235335 EEB14861.1 GFDF01003530 JAV10554.1 DS231909 EDS45626.1 KB631815 ERL86492.1 GFDL01008754 JAV26291.1 CH477280 EAT44953.1 ATLV01024424 KE525352 KFB51375.1 AXCP01009302 KA648238 AFP62867.1 JXUM01042784 KQ561338 KXJ78922.1 AXCM01003043 ADMH02001892 ETN60621.1 APCN01002331 AAAB01008960 EAA10776.1 GGMS01008364 MBY77567.1 AXCN02000395 CCAG010015406 AJWK01000520 AJWK01000521 AJWK01000522 AJWK01000523 JXJN01015978 ABLF02035837 GGMR01020067 MBY32686.1 CH916369 EDV93471.1 GDIQ01110377 JAL41349.1 GDIQ01010060 JAN84677.1 GDIP01110234 GDIP01102984 GDIQ01036907 LRGB01001574 JAL93480.1 JAN57830.1 KZS11678.1 UFQS01001174 UFQT01001174 SSX09398.1 SSX29300.1 CM000070 EAL28353.1 CH479182 EDW34046.1 GDIP01204274 JAJ19128.1 GDIP01215327 JAJ08075.1 GAMC01012882 GAMC01012880 JAB93673.1 JRES01000301 KNC32576.1 GDIP01105779 JAL97935.1 GL732531 EFX85747.1 GDHF01028125 GDHF01026495 GDHF01011125 GDHF01006198 JAI24189.1 JAI25819.1 JAI41189.1 JAI46116.1 CM000364 EDX12134.1 AE014297 BT003492 OUUW01000007 SPP83407.1 GAKP01007916 JAC51036.1 CH902617 EDV43492.1
RSAL01000054 RVE50013.1 KQ459603 KPI93105.1 AGBW02012291 OWR45272.1 KQ460711 KPJ12669.1 NEVH01019998 PNF21996.1 ACPB03019765 GEDC01009475 JAS27823.1 GEBQ01030470 JAT09507.1 GECL01002286 JAP03838.1 GBBI01002709 JAC16003.1 GBYB01000060 JAG69827.1 GBHO01039771 GBHO01022367 GBHO01022365 JAG03833.1 JAG21237.1 JAG21239.1 GL449784 EFN81987.1 GECZ01016907 JAS52862.1 KK853313 KDR08619.1 GEDC01005583 JAS31715.1 KQ434875 KZC09765.1 GEDC01027115 JAS10183.1 GEBQ01004190 JAT35787.1 GFTR01007984 JAW08442.1 LBMM01010688 KMQ87316.1 KK107295 EZA53099.1 KZ288206 PBC33071.1 GL443262 EFN62313.1 GBHO01022363 GBHO01022361 GBHO01022360 JAG21241.1 JAG21243.1 JAG21244.1 KQ414596 KOC70025.1 QOIP01000003 RLU24964.1 KQ435944 KOX68212.1 NNAY01000292 OXU29420.1 GL888280 EGI63363.1 GEZM01031081 JAV85076.1 KQ980882 KYN11946.1 ADTU01003649 ADTU01003650 KQ976709 KYM77042.1 KQ981261 KYN44139.1 KQ983089 KYQ47616.1 KQ971381 EEZ97223.1 DS235335 EEB14861.1 GFDF01003530 JAV10554.1 DS231909 EDS45626.1 KB631815 ERL86492.1 GFDL01008754 JAV26291.1 CH477280 EAT44953.1 ATLV01024424 KE525352 KFB51375.1 AXCP01009302 KA648238 AFP62867.1 JXUM01042784 KQ561338 KXJ78922.1 AXCM01003043 ADMH02001892 ETN60621.1 APCN01002331 AAAB01008960 EAA10776.1 GGMS01008364 MBY77567.1 AXCN02000395 CCAG010015406 AJWK01000520 AJWK01000521 AJWK01000522 AJWK01000523 JXJN01015978 ABLF02035837 GGMR01020067 MBY32686.1 CH916369 EDV93471.1 GDIQ01110377 JAL41349.1 GDIQ01010060 JAN84677.1 GDIP01110234 GDIP01102984 GDIQ01036907 LRGB01001574 JAL93480.1 JAN57830.1 KZS11678.1 UFQS01001174 UFQT01001174 SSX09398.1 SSX29300.1 CM000070 EAL28353.1 CH479182 EDW34046.1 GDIP01204274 JAJ19128.1 GDIP01215327 JAJ08075.1 GAMC01012882 GAMC01012880 JAB93673.1 JRES01000301 KNC32576.1 GDIP01105779 JAL97935.1 GL732531 EFX85747.1 GDHF01028125 GDHF01026495 GDHF01011125 GDHF01006198 JAI24189.1 JAI25819.1 JAI41189.1 JAI46116.1 CM000364 EDX12134.1 AE014297 BT003492 OUUW01000007 SPP83407.1 GAKP01007916 JAC51036.1 CH902617 EDV43492.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000007151
UP000053240
+ More
UP000235965 UP000015103 UP000008237 UP000027135 UP000002358 UP000076502 UP000005203 UP000036403 UP000053097 UP000242457 UP000000311 UP000053825 UP000279307 UP000053105 UP000192223 UP000215335 UP000007755 UP000078492 UP000005205 UP000078540 UP000078541 UP000075809 UP000007266 UP000009046 UP000002320 UP000030742 UP000008820 UP000030765 UP000075880 UP000076408 UP000095301 UP000069940 UP000249989 UP000076407 UP000075883 UP000075900 UP000075884 UP000075881 UP000075920 UP000000673 UP000075903 UP000075902 UP000075840 UP000069272 UP000007062 UP000075885 UP000075886 UP000095300 UP000092444 UP000092445 UP000078200 UP000092461 UP000092443 UP000092460 UP000007819 UP000091820 UP000001070 UP000076858 UP000001819 UP000008744 UP000037069 UP000000305 UP000000304 UP000000803 UP000268350 UP000007801
UP000235965 UP000015103 UP000008237 UP000027135 UP000002358 UP000076502 UP000005203 UP000036403 UP000053097 UP000242457 UP000000311 UP000053825 UP000279307 UP000053105 UP000192223 UP000215335 UP000007755 UP000078492 UP000005205 UP000078540 UP000078541 UP000075809 UP000007266 UP000009046 UP000002320 UP000030742 UP000008820 UP000030765 UP000075880 UP000076408 UP000095301 UP000069940 UP000249989 UP000076407 UP000075883 UP000075900 UP000075884 UP000075881 UP000075920 UP000000673 UP000075903 UP000075902 UP000075840 UP000069272 UP000007062 UP000075885 UP000075886 UP000095300 UP000092444 UP000092445 UP000078200 UP000092461 UP000092443 UP000092460 UP000007819 UP000091820 UP000001070 UP000076858 UP000001819 UP000008744 UP000037069 UP000000305 UP000000304 UP000000803 UP000268350 UP000007801
Pfam
PF04091 Sec15
Interpro
SUPFAM
SSF52129
SSF52129
Gene 3D
ProteinModelPortal
H9JBU2
A0A2A4JPP3
A0A2H1W243
A0A3S2M3B5
A0A194PPP6
A0A212EUV6
+ More
A0A0N1PGR5 A0A2J7Q098 T1ICC1 A0A1B6DQE0 A0A1B6KDJ2 A0A0V0G758 A0A023F4S5 A0A0C9QWG4 A0A0A9XMQ2 E2BQM2 A0A1B6FRM1 A0A067QT00 K7IYQ6 A0A1B6E1B3 A0A154PCZ2 A0A088ADK2 A0A1B6C9M8 A0A1B6MIK1 A0A224XIN4 A0A0J7KAQ1 A0A026WDL2 A0A2A3EPV3 E2AW91 A0A0A9XNN5 A0A0L7RGP6 A0A3L8DWW5 A0A0N0BC08 A0A1W4X769 A0A232FFX4 F4WR05 A0A1Y1MMF6 A0A151IVS7 A0A158NXX0 A0A151HZ51 A0A195FWC6 A0A151WIG8 D6X4X0 E0VNA5 A0A1L8DVT5 B0WEG6 U4U091 A0A1Q3FFH0 Q17EP0 A0A084WMD1 A0A1S4F5K5 A0A182J5G8 A0A182Y2G0 A0A1I8MEH2 T1PJV5 A0A182G4S8 A0A182X4M8 A0A182M0K3 A0A182RDK0 A0A182NI62 A0A182K1S6 A0A182WBM2 W5J9E9 A0A182V8N7 A0A182TGC6 A0A182I4M6 A0A182FDB4 Q7Q546 A0A2S2QIL0 A0A182PDJ7 A0A182QQG8 A0A1I8PYW4 A0A1B0G9K3 A0A1A9ZUK1 A0A1A9UZY9 A0A1B0C8E7 A0A1A9XSW6 A0A1B0BKM5 J9K5H0 A0A2S2PUZ7 A0A1A9W9E0 B4JFJ0 A0A0P5R9Z7 A0A0N8ENA1 A0A0P5USB6 A0A336KYV7 Q296V4 B4GEL1 A0A0P5AH24 A0A0P4ZLA6 W8B9F1 A0A0L0CJZ9 A0A0P5V262 E9G3C4 A0A0K8UC27 B4R147 Q9VDE6 A0A3B0JMC0 A0A034W611 B3M2W2
A0A0N1PGR5 A0A2J7Q098 T1ICC1 A0A1B6DQE0 A0A1B6KDJ2 A0A0V0G758 A0A023F4S5 A0A0C9QWG4 A0A0A9XMQ2 E2BQM2 A0A1B6FRM1 A0A067QT00 K7IYQ6 A0A1B6E1B3 A0A154PCZ2 A0A088ADK2 A0A1B6C9M8 A0A1B6MIK1 A0A224XIN4 A0A0J7KAQ1 A0A026WDL2 A0A2A3EPV3 E2AW91 A0A0A9XNN5 A0A0L7RGP6 A0A3L8DWW5 A0A0N0BC08 A0A1W4X769 A0A232FFX4 F4WR05 A0A1Y1MMF6 A0A151IVS7 A0A158NXX0 A0A151HZ51 A0A195FWC6 A0A151WIG8 D6X4X0 E0VNA5 A0A1L8DVT5 B0WEG6 U4U091 A0A1Q3FFH0 Q17EP0 A0A084WMD1 A0A1S4F5K5 A0A182J5G8 A0A182Y2G0 A0A1I8MEH2 T1PJV5 A0A182G4S8 A0A182X4M8 A0A182M0K3 A0A182RDK0 A0A182NI62 A0A182K1S6 A0A182WBM2 W5J9E9 A0A182V8N7 A0A182TGC6 A0A182I4M6 A0A182FDB4 Q7Q546 A0A2S2QIL0 A0A182PDJ7 A0A182QQG8 A0A1I8PYW4 A0A1B0G9K3 A0A1A9ZUK1 A0A1A9UZY9 A0A1B0C8E7 A0A1A9XSW6 A0A1B0BKM5 J9K5H0 A0A2S2PUZ7 A0A1A9W9E0 B4JFJ0 A0A0P5R9Z7 A0A0N8ENA1 A0A0P5USB6 A0A336KYV7 Q296V4 B4GEL1 A0A0P5AH24 A0A0P4ZLA6 W8B9F1 A0A0L0CJZ9 A0A0P5V262 E9G3C4 A0A0K8UC27 B4R147 Q9VDE6 A0A3B0JMC0 A0A034W611 B3M2W2
PDB
2A2F
E-value=8.49344e-74,
Score=707
Ontologies
GO
GO:0000145
GO:0006904
GO:0016020
GO:0006887
GO:0006893
GO:0004197
GO:0007298
GO:0022416
GO:0032456
GO:0043679
GO:0007411
GO:0017137
GO:0042331
GO:0072659
GO:0055037
GO:0016080
GO:0005768
GO:0007269
GO:0016192
GO:0016081
GO:0031410
GO:0090522
GO:0015031
GO:0008061
GO:0003743
GO:0005852
GO:0006413
GO:0031369
GO:0005515
GO:0046872
GO:0003824
PANTHER
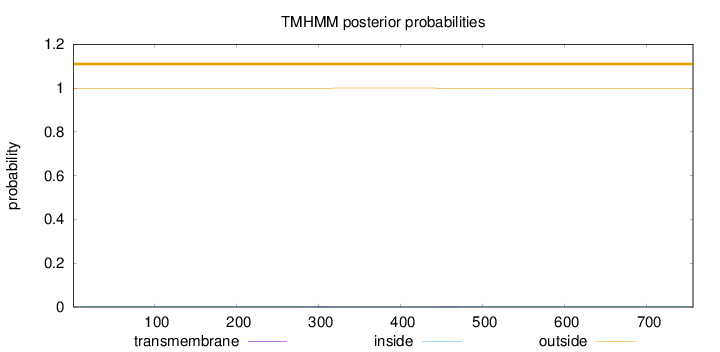
Topology
Length:
756
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0804399999999998
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00156
outside
1 - 756
Population Genetic Test Statistics
Pi
246.343436
Theta
179.327551
Tajima's D
1.227172
CLR
0.085254
CSRT
0.718514074296285
Interpretation
Uncertain
