Pre Gene Modal
BGIBMGA007119
Annotation
PREDICTED:_adenylosuccinate_lyase_[Bombyx_mori]
Full name
Adenylosuccinate lyase
Alternative Name
Adenylosuccinase
Location in the cell
Cytoplasmic Reliability : 1.656 Mitochondrial Reliability : 1.219
Sequence
CDS
ATGACCAAGATCAGTTCAACCGCAGAGCTCGGATTGGACATAAGCGCCGCTCAGATAGCCGAACTAGAAGCTGCTGTACATGACATCGACTTCCCAGCGGCTGCTGAACATGAGAAGCGGGTTCGTCACGACGTGATGGCGCATGTCCACACGCTCGGCGAGAGGTGTCCCCTCGCCGCGCCCATCATACACCTGGGAGCCACGTCGTGCTACGTCGGAGACAACACCGATTTGATAGTCCTTAAGAACGGACTCGAGCTGCTTCTGCCAAGGCTCGTCGCCGTCATCAAACAACTTGGAAGCTTCGCGAAACAGCACAAGTCTTTGCCCATCCTCGGCTTCACCCATCTTCAACCGGCGCAACTGACAACGGTCGGTAAAAGAGCATCGCTGTGGCTCAACGACTTGCTAATGGATGAACGCGCTTTGTCTCGCGCTAAAGACGACCTCCGGTTCAGGGGCGTGAAGGGTACCACGGGCACTCAGGCTTCCTTCCTCCAGCTCTTCAAAGGAGACGGTGAAAAGGTTCGCGCACTAGACAAAAGAGTTGCGGAACTGGCCGGATTTGATAAGAGATACATCGTGACCGGACAGACGTATTCCAGAAAAGTCGATTTAGAAGTTATTTCCGCATTGTCAGGCCTTGGAGCTACGGTACACAAAATGTGTTCAGACATACGTATTCTCGCCTCTCGTAAAGAGCTTGAGGAGCCTTTTGAAGTGTCCCAAATTGGTTCTAGTGCCATGCCTTACAAGAGGAACCCAATGCGTTCAGAACGTTGCTGCGCTCTAGCCCGTCATCTAATTACTTTACACGCTAACGCAGCTAATACTCATGCCGTCCAATGGTTAGAACGAACCCTAGACGACTCTGCCAATCGTCGCATTACCCTCGCCGAGGCTTTCCTTACCGCTGACGCTACTCTACTGACACTATTAAATATTTGCCAAGGCCTGGTGGTGTATCCGAAAGTGATCGCGCGTCATATCGCTCAAGAACTACCTTTCATGGCGACGGAAAATATAATAATGGCTATGGTCCAAGCGGGAGGTGACCGTCAAGTGTGCCACGAAAAGATTAGGGTGCTCTCCCATGAAGCAGGGGCTCAAGTGAAGCAACACGGCAGAGATAATGATTTGATTGAACGCGTTAAGAAAGACGGCTATTTCGCCCCAATTATATCCCAGTTGGACAAGATATTGGACGCTTCGACGTTCATTGGTAGAGCTCCAGAACAAGTGGACGAGTTTTTGGATGAAGAAGTCGATCCCATTATCGCTAAATACGCAGATTCCCTTATAAATGTTGACAAGCCAGTGGCACTAAACATTTAA
Protein
MTKISSTAELGLDISAAQIAELEAAVHDIDFPAAAEHEKRVRHDVMAHVHTLGERCPLAAPIIHLGATSCYVGDNTDLIVLKNGLELLLPRLVAVIKQLGSFAKQHKSLPILGFTHLQPAQLTTVGKRASLWLNDLLMDERALSRAKDDLRFRGVKGTTGTQASFLQLFKGDGEKVRALDKRVAELAGFDKRYIVTGQTYSRKVDLEVISALSGLGATVHKMCSDIRILASRKELEEPFEVSQIGSSAMPYKRNPMRSERCCALARHLITLHANAANTHAVQWLERTLDDSANRRITLAEAFLTADATLLTLLNICQGLVVYPKVIARHIAQELPFMATENIIMAMVQAGGDRQVCHEKIRVLSHEAGAQVKQHGRDNDLIERVKKDGYFAPIISQLDKILDASTFIGRAPEQVDEFLDEEVDPIIAKYADSLINVDKPVALNI
Summary
Catalytic Activity
(2S)-2-[5-amino-1-(5-phospho-beta-D-ribosyl)imidazole-4-carboxamido]succinate = 5-amino-1-(5-phospho-beta-D-ribosyl)imidazole-4-carboxamide + fumarate
N(6)-(1,2-dicarboxyethyl)-AMP = AMP + fumarate
N(6)-(1,2-dicarboxyethyl)-AMP = AMP + fumarate
Similarity
Belongs to the lyase 1 family. Adenylosuccinate lyase subfamily.
Uniprot
H9JC74
A0A0N1PHN5
A0A2H1V6T9
B2DBN3
A0A212EUV7
A0A2A4JHM0
+ More
A0A3S2NFU5 A0A0L7LGT3 S4PF95 A0A194PIF9 A0A182GJX8 A0A1L8DXT9 A0A023ESY0 B4PM94 A0A1Q3F8X8 J9HT71 A0A1B0CJJ2 J9HJI9 A0A3B0JLJ4 A0A182HT94 B0W6F1 Q7QBZ6 A0A182K1Z5 B4NJF6 B3P3J2 A0A182XIA7 A0A182V252 B4QWD5 A0A182PA48 A0A182U1L3 A0A1W4X215 Q9VEP6 A0A1W4VH91 A0A182IWI7 B4IBN8 A0A0M5J4C2 B4KV48 Q95SP9 B3M2B5 A0A182NA73 W5JM61 A0A182F8B9 A0A182QYL4 A0A2M4ACA5 A0A2M4BLZ9 A0A2M4BLP6 B4IYT5 A0A2M4BLM0 A0A2M3Z1C5 B4LDA2 A0A182LVZ8 A0A182YS80 A0A182W2W6 U5EUN6 A0A182RYG0 B4G4K3 Q297I4 A0A0L0CRA3 K7IMY2 T1PGQ8 A0A1I8P9R0 A0A0T6AUS5 A0A1Y1LA35 E2ASU4 A0A0K8TL45 A0A0A1X0W1 A0A084W1M2 A0A336KCM1 A0A026X3L2 W8C837 A0A0K8UL87 A0A034WS39 A0A0L7QQX1 A0A0M8ZUQ3 D6WX11 E2BFA3 A0A151IU78 A0A154P2M6 A0A151IAQ0 F4X2U5 A0A151WKK4 A0A2A3EC47 A0A158NLQ0 A0A195BLA0 A0A195FVD7 A0A232F4U8 A0A0C9PVA4 U4TSL0 A0A1A9W589 A0A1B0AWJ6 A0A3L8DIQ7 A0A0K8V1A4 A0A1A9Z7E5 D3TS08 A0A1A9Y5Y6 A0A1A9V512 E9J7N7 A0A1J1HY75 A0A087ZXK2 T1DST8
A0A3S2NFU5 A0A0L7LGT3 S4PF95 A0A194PIF9 A0A182GJX8 A0A1L8DXT9 A0A023ESY0 B4PM94 A0A1Q3F8X8 J9HT71 A0A1B0CJJ2 J9HJI9 A0A3B0JLJ4 A0A182HT94 B0W6F1 Q7QBZ6 A0A182K1Z5 B4NJF6 B3P3J2 A0A182XIA7 A0A182V252 B4QWD5 A0A182PA48 A0A182U1L3 A0A1W4X215 Q9VEP6 A0A1W4VH91 A0A182IWI7 B4IBN8 A0A0M5J4C2 B4KV48 Q95SP9 B3M2B5 A0A182NA73 W5JM61 A0A182F8B9 A0A182QYL4 A0A2M4ACA5 A0A2M4BLZ9 A0A2M4BLP6 B4IYT5 A0A2M4BLM0 A0A2M3Z1C5 B4LDA2 A0A182LVZ8 A0A182YS80 A0A182W2W6 U5EUN6 A0A182RYG0 B4G4K3 Q297I4 A0A0L0CRA3 K7IMY2 T1PGQ8 A0A1I8P9R0 A0A0T6AUS5 A0A1Y1LA35 E2ASU4 A0A0K8TL45 A0A0A1X0W1 A0A084W1M2 A0A336KCM1 A0A026X3L2 W8C837 A0A0K8UL87 A0A034WS39 A0A0L7QQX1 A0A0M8ZUQ3 D6WX11 E2BFA3 A0A151IU78 A0A154P2M6 A0A151IAQ0 F4X2U5 A0A151WKK4 A0A2A3EC47 A0A158NLQ0 A0A195BLA0 A0A195FVD7 A0A232F4U8 A0A0C9PVA4 U4TSL0 A0A1A9W589 A0A1B0AWJ6 A0A3L8DIQ7 A0A0K8V1A4 A0A1A9Z7E5 D3TS08 A0A1A9Y5Y6 A0A1A9V512 E9J7N7 A0A1J1HY75 A0A087ZXK2 T1DST8
EC Number
4.3.2.2
Pubmed
19121390
26354079
18712529
22118469
26227816
23622113
+ More
26483478 24945155 17994087 17550304 17510324 12364791 14747013 17210077 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20920257 23761445 25244985 15632085 26108605 20075255 25315136 28004739 20798317 26369729 25830018 24438588 24508170 24495485 25348373 18362917 19820115 21719571 21347285 28648823 23537049 30249741 20353571 21282665
26483478 24945155 17994087 17550304 17510324 12364791 14747013 17210077 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20920257 23761445 25244985 15632085 26108605 20075255 25315136 28004739 20798317 26369729 25830018 24438588 24508170 24495485 25348373 18362917 19820115 21719571 21347285 28648823 23537049 30249741 20353571 21282665
EMBL
BABH01015027
KQ460711
KPJ12664.1
ODYU01000980
SOQ36548.1
AB264718
+ More
BAG30792.1 AGBW02012291 OWR45275.1 NWSH01001534 PCG70933.1 RSAL01000054 RVE50016.1 JTDY01001181 KOB74630.1 GAIX01004132 JAA88428.1 KQ459603 KPI93102.1 JXUM01013063 KQ560367 KXJ82788.1 GFDF01002823 JAV11261.1 GAPW01001183 JAC12415.1 CM000160 EDW96960.1 GFDL01011021 JAV24024.1 CH477623 EJY57843.1 AJWK01014673 CH478271 EJY58099.1 OUUW01000007 SPP83194.1 APCN01000423 DS231848 EDS36625.1 AAAB01008859 EAA07522.3 CH964272 EDW83880.1 CH954181 EDV48772.1 CM000364 EDX12626.1 AE014297 BT044196 AAF55375.1 ACH92261.1 CH480827 EDW44796.1 CP012525 ALC45211.1 CH933809 EDW19388.1 AY060661 AAL28209.1 CH902617 EDV42306.1 ADMH02000650 ETN65442.1 AXCN02000218 GGFK01005092 MBW38413.1 GGFJ01004830 MBW53971.1 GGFJ01004834 MBW53975.1 CH916366 EDV96622.1 GGFJ01004835 MBW53976.1 GGFM01001579 MBW22330.1 CH940647 EDW71009.1 AXCM01000066 GANO01002237 JAB57634.1 CH479179 EDW25174.1 CM000070 EAL28221.1 JRES01000032 KNC34766.1 KA647335 AFP61964.1 LJIG01022818 KRT78558.1 GEZM01061414 JAV70504.1 GL442359 EFN63519.1 GDAI01002539 JAI15064.1 GBXI01010029 GBXI01006344 JAD04263.1 JAD07948.1 ATLV01019388 KE525269 KFB44116.1 UFQS01000302 UFQT01000302 SSX02646.1 SSX23020.1 KK107019 EZA62653.1 GAMC01003244 JAC03312.1 GDHF01024998 GDHF01020518 JAI27316.1 JAI31796.1 GAKP01001947 JAC57005.1 KQ414784 KOC61028.1 KQ435834 KOX71530.1 KQ971361 EFA08776.1 GL447982 EFN85616.1 KQ980968 KYN10973.1 KQ434796 KZC05594.1 KQ978186 KYM96559.1 GL888591 EGI59291.1 KQ983020 KYQ48215.1 KZ288288 PBC29353.1 ADTU01019629 ADTU01019630 ADTU01019631 ADTU01019632 KQ976450 KYM85641.1 KQ981219 KYN44391.1 NNAY01000965 OXU25705.1 GBYB01005338 JAG75105.1 KB631584 ERL84499.1 JXJN01004810 QOIP01000007 RLU20335.1 GDHF01019607 GDHF01007503 JAI32707.1 JAI44811.1 CCAG010018629 EZ424210 ADD20486.1 GL768590 EFZ11155.1 CVRI01000021 CRK91486.1 GAMD01001334 JAB00257.1
BAG30792.1 AGBW02012291 OWR45275.1 NWSH01001534 PCG70933.1 RSAL01000054 RVE50016.1 JTDY01001181 KOB74630.1 GAIX01004132 JAA88428.1 KQ459603 KPI93102.1 JXUM01013063 KQ560367 KXJ82788.1 GFDF01002823 JAV11261.1 GAPW01001183 JAC12415.1 CM000160 EDW96960.1 GFDL01011021 JAV24024.1 CH477623 EJY57843.1 AJWK01014673 CH478271 EJY58099.1 OUUW01000007 SPP83194.1 APCN01000423 DS231848 EDS36625.1 AAAB01008859 EAA07522.3 CH964272 EDW83880.1 CH954181 EDV48772.1 CM000364 EDX12626.1 AE014297 BT044196 AAF55375.1 ACH92261.1 CH480827 EDW44796.1 CP012525 ALC45211.1 CH933809 EDW19388.1 AY060661 AAL28209.1 CH902617 EDV42306.1 ADMH02000650 ETN65442.1 AXCN02000218 GGFK01005092 MBW38413.1 GGFJ01004830 MBW53971.1 GGFJ01004834 MBW53975.1 CH916366 EDV96622.1 GGFJ01004835 MBW53976.1 GGFM01001579 MBW22330.1 CH940647 EDW71009.1 AXCM01000066 GANO01002237 JAB57634.1 CH479179 EDW25174.1 CM000070 EAL28221.1 JRES01000032 KNC34766.1 KA647335 AFP61964.1 LJIG01022818 KRT78558.1 GEZM01061414 JAV70504.1 GL442359 EFN63519.1 GDAI01002539 JAI15064.1 GBXI01010029 GBXI01006344 JAD04263.1 JAD07948.1 ATLV01019388 KE525269 KFB44116.1 UFQS01000302 UFQT01000302 SSX02646.1 SSX23020.1 KK107019 EZA62653.1 GAMC01003244 JAC03312.1 GDHF01024998 GDHF01020518 JAI27316.1 JAI31796.1 GAKP01001947 JAC57005.1 KQ414784 KOC61028.1 KQ435834 KOX71530.1 KQ971361 EFA08776.1 GL447982 EFN85616.1 KQ980968 KYN10973.1 KQ434796 KZC05594.1 KQ978186 KYM96559.1 GL888591 EGI59291.1 KQ983020 KYQ48215.1 KZ288288 PBC29353.1 ADTU01019629 ADTU01019630 ADTU01019631 ADTU01019632 KQ976450 KYM85641.1 KQ981219 KYN44391.1 NNAY01000965 OXU25705.1 GBYB01005338 JAG75105.1 KB631584 ERL84499.1 JXJN01004810 QOIP01000007 RLU20335.1 GDHF01019607 GDHF01007503 JAI32707.1 JAI44811.1 CCAG010018629 EZ424210 ADD20486.1 GL768590 EFZ11155.1 CVRI01000021 CRK91486.1 GAMD01001334 JAB00257.1
Proteomes
UP000005204
UP000053240
UP000007151
UP000218220
UP000283053
UP000037510
+ More
UP000053268 UP000069940 UP000249989 UP000002282 UP000008820 UP000092461 UP000268350 UP000075840 UP000002320 UP000007062 UP000075881 UP000007798 UP000008711 UP000076407 UP000075903 UP000000304 UP000075885 UP000075902 UP000192223 UP000000803 UP000192221 UP000075880 UP000001292 UP000092553 UP000009192 UP000007801 UP000075884 UP000000673 UP000069272 UP000075886 UP000001070 UP000008792 UP000075883 UP000076408 UP000075920 UP000075900 UP000008744 UP000001819 UP000037069 UP000002358 UP000095301 UP000095300 UP000000311 UP000030765 UP000053097 UP000053825 UP000053105 UP000007266 UP000008237 UP000078492 UP000076502 UP000078542 UP000007755 UP000075809 UP000242457 UP000005205 UP000078540 UP000078541 UP000215335 UP000030742 UP000091820 UP000092460 UP000279307 UP000092445 UP000092444 UP000092443 UP000078200 UP000183832 UP000005203
UP000053268 UP000069940 UP000249989 UP000002282 UP000008820 UP000092461 UP000268350 UP000075840 UP000002320 UP000007062 UP000075881 UP000007798 UP000008711 UP000076407 UP000075903 UP000000304 UP000075885 UP000075902 UP000192223 UP000000803 UP000192221 UP000075880 UP000001292 UP000092553 UP000009192 UP000007801 UP000075884 UP000000673 UP000069272 UP000075886 UP000001070 UP000008792 UP000075883 UP000076408 UP000075920 UP000075900 UP000008744 UP000001819 UP000037069 UP000002358 UP000095301 UP000095300 UP000000311 UP000030765 UP000053097 UP000053825 UP000053105 UP000007266 UP000008237 UP000078492 UP000076502 UP000078542 UP000007755 UP000075809 UP000242457 UP000005205 UP000078540 UP000078541 UP000215335 UP000030742 UP000091820 UP000092460 UP000279307 UP000092445 UP000092444 UP000092443 UP000078200 UP000183832 UP000005203
Interpro
IPR022761
Fumarate_lyase_N
+ More
IPR020557 Fumarate_lyase_CS
IPR019468 AdenyloSucc_lyase_C
IPR000362 Fumarate_lyase_fam
IPR008948 L-Aspartase-like
IPR004769 Pur_lyase
IPR024083 Fumarase/histidase_N
IPR001995 Peptidase_A2_cat
IPR021109 Peptidase_aspartic_dom_sf
IPR035979 RBD_domain_sf
IPR000504 RRM_dom
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR020557 Fumarate_lyase_CS
IPR019468 AdenyloSucc_lyase_C
IPR000362 Fumarate_lyase_fam
IPR008948 L-Aspartase-like
IPR004769 Pur_lyase
IPR024083 Fumarase/histidase_N
IPR001995 Peptidase_A2_cat
IPR021109 Peptidase_aspartic_dom_sf
IPR035979 RBD_domain_sf
IPR000504 RRM_dom
IPR012677 Nucleotide-bd_a/b_plait_sf
Gene 3D
ProteinModelPortal
H9JC74
A0A0N1PHN5
A0A2H1V6T9
B2DBN3
A0A212EUV7
A0A2A4JHM0
+ More
A0A3S2NFU5 A0A0L7LGT3 S4PF95 A0A194PIF9 A0A182GJX8 A0A1L8DXT9 A0A023ESY0 B4PM94 A0A1Q3F8X8 J9HT71 A0A1B0CJJ2 J9HJI9 A0A3B0JLJ4 A0A182HT94 B0W6F1 Q7QBZ6 A0A182K1Z5 B4NJF6 B3P3J2 A0A182XIA7 A0A182V252 B4QWD5 A0A182PA48 A0A182U1L3 A0A1W4X215 Q9VEP6 A0A1W4VH91 A0A182IWI7 B4IBN8 A0A0M5J4C2 B4KV48 Q95SP9 B3M2B5 A0A182NA73 W5JM61 A0A182F8B9 A0A182QYL4 A0A2M4ACA5 A0A2M4BLZ9 A0A2M4BLP6 B4IYT5 A0A2M4BLM0 A0A2M3Z1C5 B4LDA2 A0A182LVZ8 A0A182YS80 A0A182W2W6 U5EUN6 A0A182RYG0 B4G4K3 Q297I4 A0A0L0CRA3 K7IMY2 T1PGQ8 A0A1I8P9R0 A0A0T6AUS5 A0A1Y1LA35 E2ASU4 A0A0K8TL45 A0A0A1X0W1 A0A084W1M2 A0A336KCM1 A0A026X3L2 W8C837 A0A0K8UL87 A0A034WS39 A0A0L7QQX1 A0A0M8ZUQ3 D6WX11 E2BFA3 A0A151IU78 A0A154P2M6 A0A151IAQ0 F4X2U5 A0A151WKK4 A0A2A3EC47 A0A158NLQ0 A0A195BLA0 A0A195FVD7 A0A232F4U8 A0A0C9PVA4 U4TSL0 A0A1A9W589 A0A1B0AWJ6 A0A3L8DIQ7 A0A0K8V1A4 A0A1A9Z7E5 D3TS08 A0A1A9Y5Y6 A0A1A9V512 E9J7N7 A0A1J1HY75 A0A087ZXK2 T1DST8
A0A3S2NFU5 A0A0L7LGT3 S4PF95 A0A194PIF9 A0A182GJX8 A0A1L8DXT9 A0A023ESY0 B4PM94 A0A1Q3F8X8 J9HT71 A0A1B0CJJ2 J9HJI9 A0A3B0JLJ4 A0A182HT94 B0W6F1 Q7QBZ6 A0A182K1Z5 B4NJF6 B3P3J2 A0A182XIA7 A0A182V252 B4QWD5 A0A182PA48 A0A182U1L3 A0A1W4X215 Q9VEP6 A0A1W4VH91 A0A182IWI7 B4IBN8 A0A0M5J4C2 B4KV48 Q95SP9 B3M2B5 A0A182NA73 W5JM61 A0A182F8B9 A0A182QYL4 A0A2M4ACA5 A0A2M4BLZ9 A0A2M4BLP6 B4IYT5 A0A2M4BLM0 A0A2M3Z1C5 B4LDA2 A0A182LVZ8 A0A182YS80 A0A182W2W6 U5EUN6 A0A182RYG0 B4G4K3 Q297I4 A0A0L0CRA3 K7IMY2 T1PGQ8 A0A1I8P9R0 A0A0T6AUS5 A0A1Y1LA35 E2ASU4 A0A0K8TL45 A0A0A1X0W1 A0A084W1M2 A0A336KCM1 A0A026X3L2 W8C837 A0A0K8UL87 A0A034WS39 A0A0L7QQX1 A0A0M8ZUQ3 D6WX11 E2BFA3 A0A151IU78 A0A154P2M6 A0A151IAQ0 F4X2U5 A0A151WKK4 A0A2A3EC47 A0A158NLQ0 A0A195BLA0 A0A195FVD7 A0A232F4U8 A0A0C9PVA4 U4TSL0 A0A1A9W589 A0A1B0AWJ6 A0A3L8DIQ7 A0A0K8V1A4 A0A1A9Z7E5 D3TS08 A0A1A9Y5Y6 A0A1A9V512 E9J7N7 A0A1J1HY75 A0A087ZXK2 T1DST8
PDB
2VD6
E-value=5.10229e-164,
Score=1483
Ontologies
PATHWAY
GO
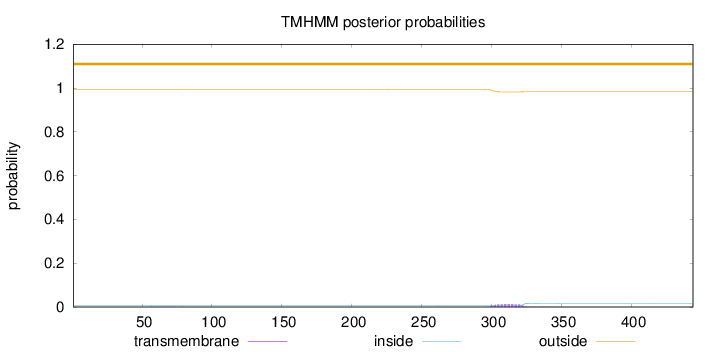
Topology
Length:
444
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.28763
Exp number, first 60 AAs:
0.00119
Total prob of N-in:
0.00754
outside
1 - 444
Population Genetic Test Statistics
Pi
180.014273
Theta
160.298086
Tajima's D
1.510844
CLR
0.012015
CSRT
0.790460476976151
Interpretation
Uncertain
