Gene
KWMTBOMO10499
Pre Gene Modal
BGIBMGA006990
Annotation
ryanodine_receptor-like_protein_[Danaus_plexippus]
Full name
Protein snakeskin
Location in the cell
PlasmaMembrane Reliability : 4.351
Sequence
CDS
ATGGTTTCCGTTCAGACTATAGCGACTATAGTCGTGAAGACATTCAAAATTGTCTTGAACATTATAATCTTAGTGCTCTACCGCACCGGTTACAATGGAGAGTTCCTGGGGGTCGGAGGAACTTGGAACCTGAACGAGGAGAAGAATCCTGACGCGGAGATCGTTGCCTCAGGAGTAATCGTGGGCTACCTCATTTATACGCTAGTGCAGATCGTCACATTCTTGTTTGGTACCACGGAGCACAAACGCGCTTTGTCTGAGATCGTGATGAACTTCGTCGGAGTGTTCCTATGGATTGCGGTGGGCGCTGTGGCATTACACTACTGGGGAGGATACCAGGGCGAGCACCAGTTCCAGTTTGTCTTCGCTGAGAAACAGGTTGGCCTGGCCGTCGGAGCTCTATGCGTCATAAATGGAGCCATATATTTATTGGACACGGCACTTTCTGTCATACATTTCACAAAGGAAATGTAA
Protein
MVSVQTIATIVVKTFKIVLNIIILVLYRTGYNGEFLGVGGTWNLNEEKNPDAEIVASGVIVGYLIYTLVQIVTFLFGTTEHKRALSEIVMNFVGVFLWIAVGAVALHYWGGYQGEHQFQFVFAEKQVGLAVGALCVINGAIYLLDTALSVIHFTKEM
Summary
Description
Required for assembly of smooth septate junctions (sSJs). May be important for barrier function of the midgut epithelium.
Required for assembly of smooth septate junctions (sSJs), together with mesh and Tsp2A (PubMed:22854041, PubMed:26848177). May be important for barrier function of the midgut epithelium.
Required for assembly of smooth septate junctions (sSJs), together with mesh and Tsp2A (PubMed:22854041, PubMed:26848177). May be important for barrier function of the midgut epithelium.
Subunit
Forms a complex with Tsp2A and mesh (PubMed:26848177). Interacts with mesh; the interaction may be necessary for the localization of both proteins to the cell apicolateral region (PubMed:22854041).
Keywords
Cell junction
Cell membrane
Complete proteome
Direct protein sequencing
Membrane
Reference proteome
Transmembrane
Transmembrane helix
Feature
chain Protein snakeskin
Uniprot
H9JBU5
A0A2H1V6X4
A0A2A4JGU3
A0A212EUW1
A0A1D5RMU7
A0A194PI55
+ More
A8C9Z3 A0A1L8DJL8 A0A1B0GL15 E9J7N8 A0A1Q3FMB6 A0A1Q3FLV3 A0A1W4XN22 A0A1B6HMV8 A0A1B6L527 A0A1B6FGC7 A0A0L7LGW8 A0A067QYM7 T1PAC6 A0A1B6ELH2 A0A182KCR6 A0A1B0G911 A0A1A9Y043 A0A1A9UQR6 A0A1A9Z2Y7 A0A1A9WRH7 A0A2J7QHB5 A0A0C9R279 A0A182IQH7 A0A0A1XJB2 A0A1B1ZGG5 A0A182QIU4 A0A182LUW7 A0A182VLD6 Q7QDX7 A0A182HXD7 A0A182LND7 A0A182YIT5 A0A182FBV4 A0A084VUP8 T1DKF2 W8C7U8 E2ASU3 A0A182RJS8 W5JJV6 B4MKW3 A0A1Y1LLN1 B4LG14 B3NE42 B4IYP9 A0A182NDF8 B4KV85 A0A0J7K5N8 Q17HS5 A0A182PLU4 A0A1W4VU80 B4PFD8 A0APU7 C0MKE7 Q9VW87 A0A182X1N6 A0A3B0JRD9 A0A182W3X0 A0A182SUE3 A0A026X5F3 A0A1J1IGG0 B4IA52 A0A195BLG1 A0A158NLP9 A0A1S4F265 B3M4P7 A0A232F5E7 B4ITJ5 Q2M0X4 B4GQZ0 B0WA98 C4WUQ0 K7JHR0 A0A087ZXG6 A0A151WK34 A0A2P8ZPA6 A0A195FVQ8 A0A336M3J5 A0A2S2PL14 J3JYN4 A0A170YLU5 D1GZ07 D1GYQ4 A0A151IAM1 A0A1B6D7J7 A0A1B0B3K6 A0A0L7QR41 A0A1L8EHS0 A0A0L0C0F2 D6WX16 A0A0K8UXH7 A0A034WLF9 A0A3Q0IZ88 A0A146M3W1
A8C9Z3 A0A1L8DJL8 A0A1B0GL15 E9J7N8 A0A1Q3FMB6 A0A1Q3FLV3 A0A1W4XN22 A0A1B6HMV8 A0A1B6L527 A0A1B6FGC7 A0A0L7LGW8 A0A067QYM7 T1PAC6 A0A1B6ELH2 A0A182KCR6 A0A1B0G911 A0A1A9Y043 A0A1A9UQR6 A0A1A9Z2Y7 A0A1A9WRH7 A0A2J7QHB5 A0A0C9R279 A0A182IQH7 A0A0A1XJB2 A0A1B1ZGG5 A0A182QIU4 A0A182LUW7 A0A182VLD6 Q7QDX7 A0A182HXD7 A0A182LND7 A0A182YIT5 A0A182FBV4 A0A084VUP8 T1DKF2 W8C7U8 E2ASU3 A0A182RJS8 W5JJV6 B4MKW3 A0A1Y1LLN1 B4LG14 B3NE42 B4IYP9 A0A182NDF8 B4KV85 A0A0J7K5N8 Q17HS5 A0A182PLU4 A0A1W4VU80 B4PFD8 A0APU7 C0MKE7 Q9VW87 A0A182X1N6 A0A3B0JRD9 A0A182W3X0 A0A182SUE3 A0A026X5F3 A0A1J1IGG0 B4IA52 A0A195BLG1 A0A158NLP9 A0A1S4F265 B3M4P7 A0A232F5E7 B4ITJ5 Q2M0X4 B4GQZ0 B0WA98 C4WUQ0 K7JHR0 A0A087ZXG6 A0A151WK34 A0A2P8ZPA6 A0A195FVQ8 A0A336M3J5 A0A2S2PL14 J3JYN4 A0A170YLU5 D1GZ07 D1GYQ4 A0A151IAM1 A0A1B6D7J7 A0A1B0B3K6 A0A0L7QR41 A0A1L8EHS0 A0A0L0C0F2 D6WX16 A0A0K8UXH7 A0A034WLF9 A0A3Q0IZ88 A0A146M3W1
Pubmed
22328496
15591204
22118469
19121390
26354079
17760985
+ More
21282665 26227816 24845553 25315136 25830018 27464714 12364791 14747013 17210077 20966253 25244985 24438588 24495485 20798317 20920257 23761445 17994087 18057021 28004739 17510324 17550304 16951084 22936249 19126864 10731132 12537572 12537569 22854041 26848177 24508170 30249741 21347285 28648823 15632085 20075255 29403074 22516182 26108605 18362917 19820115 25348373 26823975
21282665 26227816 24845553 25315136 25830018 27464714 12364791 14747013 17210077 20966253 25244985 24438588 24495485 20798317 20920257 23761445 17994087 18057021 28004739 17510324 17550304 16951084 22936249 19126864 10731132 12537572 12537569 22854041 26848177 24508170 30249741 21347285 28648823 15632085 20075255 29403074 22516182 26108605 18362917 19820115 25348373 26823975
EMBL
BABH01015024
ODYU01000980
SOQ36549.1
NWSH01001534
PCG70938.1
AGBW02012291
+ More
OWR45276.1 KQ459603 KPI93101.1 AJVK01015692 EU035817 ABV44719.1 GFDF01007438 JAV06646.1 AJWK01034052 GL768590 EFZ11159.1 GFDL01006402 JAV28643.1 GFDL01006445 JAV28600.1 GECU01031702 JAS76004.1 GEBQ01021246 GEBQ01008502 JAT18731.1 JAT31475.1 GECZ01029451 GECZ01027120 GECZ01021632 GECZ01020507 GECZ01002767 JAS40318.1 JAS42649.1 JAS48137.1 JAS49262.1 JAS67002.1 JTDY01001181 KOB74629.1 KK853186 KDR10074.1 KA645687 AFP60316.1 GECZ01031026 JAS38743.1 CCAG010002958 NEVH01013984 PNF27966.1 GBYB01006942 JAG76709.1 GBXI01003250 JAD11042.1 KU562965 ANX99820.1 AXCN02002123 AXCM01000309 AAAB01008849 EAA07189.3 APCN01005181 ATLV01016979 KE525139 KFB41692.1 GAMD01000961 JAB00630.1 GAMC01000317 JAC06239.1 GL442359 EFN63518.1 ADMH02001310 ETN63059.1 CH963847 EDW72888.1 KRF97650.1 GEZM01056124 JAV72835.1 CH940647 EDW70413.1 CH954178 EDV52466.1 CH916366 EDV96586.1 CH933809 EDW19425.1 LBMM01013146 KMQ85798.1 CH477246 EAT46164.1 CM000159 EDW95224.1 AM294685 CM000363 CM002912 CAL26671.1 EDX11186.1 KMZ00695.1 AM294673 AM294674 AM294675 AM294676 AM294677 AM294678 AM294679 AM294680 AM294681 AM294682 AM294683 AM294684 FM245818 FM245819 FM245820 FM245821 FM245822 FM245823 FM245824 FM245825 FM245826 FM245827 FM245828 FM245829 CAL26659.1 CAL26660.1 CAL26661.1 CAL26662.1 CAL26663.1 CAL26664.1 CAL26665.1 CAL26666.1 CAL26667.1 CAL26668.1 CAL26669.1 CAL26670.1 CAR93744.1 CAR93745.1 CAR93746.1 CAR93747.1 CAR93748.1 CAR93749.1 CAR93750.1 CAR93751.1 CAR93752.1 CAR93753.1 CAR93754.1 CAR93755.1 AE014296 AY052037 BT122195 AAK93461.1 ADE60669.1 OUUW01000009 SPP84704.1 KK107019 QOIP01000007 EZA62654.1 RLU20887.1 CVRI01000048 CRK98858.1 CH480826 EDW44165.1 KQ976450 KYM85642.1 ADTU01019629 CH902618 EDV39446.1 NNAY01000965 OXU25700.1 CH891711 EDW99708.1 CH379069 EAL30802.1 CH479188 EDW40175.1 DS231870 EDS40988.1 ABLF02037977 AK341164 BAH71620.1 KQ983020 KYQ48218.1 PYGN01000005 PSN58310.1 KQ981219 KYN44392.1 UFQS01000099 UFQS01000360 UFQT01000099 UFQT01000360 SSW99469.1 SSX23503.1 GGMR01017532 MBY30151.1 BT128363 AEE63321.1 GEMB01003174 JAS00037.1 FN546746 CBE67045.1 FN546643 FN546644 FN546645 FN546646 FN546647 FN546648 FN546649 FN546650 FN546651 FN546652 FN546653 FN546654 CBE66942.1 CBE66943.1 CBE66944.1 CBE66945.1 CBE66946.1 CBE66947.1 CBE66948.1 CBE66949.1 CBE66950.1 CBE66951.1 CBE66952.1 CBE66953.1 KQ978186 KYM96560.1 GEDC01025853 GEDC01015753 JAS11445.1 JAS21545.1 JXJN01007886 KQ414784 KOC61029.1 GFDG01000594 JAV18205.1 JRES01001072 KNC25778.1 KQ971361 EFA08057.1 GDHF01020937 JAI31377.1 GAKP01003967 GAKP01003966 JAC54985.1 GDHC01004674 JAQ13955.1
OWR45276.1 KQ459603 KPI93101.1 AJVK01015692 EU035817 ABV44719.1 GFDF01007438 JAV06646.1 AJWK01034052 GL768590 EFZ11159.1 GFDL01006402 JAV28643.1 GFDL01006445 JAV28600.1 GECU01031702 JAS76004.1 GEBQ01021246 GEBQ01008502 JAT18731.1 JAT31475.1 GECZ01029451 GECZ01027120 GECZ01021632 GECZ01020507 GECZ01002767 JAS40318.1 JAS42649.1 JAS48137.1 JAS49262.1 JAS67002.1 JTDY01001181 KOB74629.1 KK853186 KDR10074.1 KA645687 AFP60316.1 GECZ01031026 JAS38743.1 CCAG010002958 NEVH01013984 PNF27966.1 GBYB01006942 JAG76709.1 GBXI01003250 JAD11042.1 KU562965 ANX99820.1 AXCN02002123 AXCM01000309 AAAB01008849 EAA07189.3 APCN01005181 ATLV01016979 KE525139 KFB41692.1 GAMD01000961 JAB00630.1 GAMC01000317 JAC06239.1 GL442359 EFN63518.1 ADMH02001310 ETN63059.1 CH963847 EDW72888.1 KRF97650.1 GEZM01056124 JAV72835.1 CH940647 EDW70413.1 CH954178 EDV52466.1 CH916366 EDV96586.1 CH933809 EDW19425.1 LBMM01013146 KMQ85798.1 CH477246 EAT46164.1 CM000159 EDW95224.1 AM294685 CM000363 CM002912 CAL26671.1 EDX11186.1 KMZ00695.1 AM294673 AM294674 AM294675 AM294676 AM294677 AM294678 AM294679 AM294680 AM294681 AM294682 AM294683 AM294684 FM245818 FM245819 FM245820 FM245821 FM245822 FM245823 FM245824 FM245825 FM245826 FM245827 FM245828 FM245829 CAL26659.1 CAL26660.1 CAL26661.1 CAL26662.1 CAL26663.1 CAL26664.1 CAL26665.1 CAL26666.1 CAL26667.1 CAL26668.1 CAL26669.1 CAL26670.1 CAR93744.1 CAR93745.1 CAR93746.1 CAR93747.1 CAR93748.1 CAR93749.1 CAR93750.1 CAR93751.1 CAR93752.1 CAR93753.1 CAR93754.1 CAR93755.1 AE014296 AY052037 BT122195 AAK93461.1 ADE60669.1 OUUW01000009 SPP84704.1 KK107019 QOIP01000007 EZA62654.1 RLU20887.1 CVRI01000048 CRK98858.1 CH480826 EDW44165.1 KQ976450 KYM85642.1 ADTU01019629 CH902618 EDV39446.1 NNAY01000965 OXU25700.1 CH891711 EDW99708.1 CH379069 EAL30802.1 CH479188 EDW40175.1 DS231870 EDS40988.1 ABLF02037977 AK341164 BAH71620.1 KQ983020 KYQ48218.1 PYGN01000005 PSN58310.1 KQ981219 KYN44392.1 UFQS01000099 UFQS01000360 UFQT01000099 UFQT01000360 SSW99469.1 SSX23503.1 GGMR01017532 MBY30151.1 BT128363 AEE63321.1 GEMB01003174 JAS00037.1 FN546746 CBE67045.1 FN546643 FN546644 FN546645 FN546646 FN546647 FN546648 FN546649 FN546650 FN546651 FN546652 FN546653 FN546654 CBE66942.1 CBE66943.1 CBE66944.1 CBE66945.1 CBE66946.1 CBE66947.1 CBE66948.1 CBE66949.1 CBE66950.1 CBE66951.1 CBE66952.1 CBE66953.1 KQ978186 KYM96560.1 GEDC01025853 GEDC01015753 JAS11445.1 JAS21545.1 JXJN01007886 KQ414784 KOC61029.1 GFDG01000594 JAV18205.1 JRES01001072 KNC25778.1 KQ971361 EFA08057.1 GDHF01020937 JAI31377.1 GAKP01003967 GAKP01003966 JAC54985.1 GDHC01004674 JAQ13955.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000092462
UP000092461
+ More
UP000192223 UP000037510 UP000027135 UP000095301 UP000075881 UP000092444 UP000092443 UP000078200 UP000092445 UP000091820 UP000235965 UP000075880 UP000075886 UP000075883 UP000075903 UP000007062 UP000075840 UP000075882 UP000076408 UP000069272 UP000030765 UP000000311 UP000075900 UP000000673 UP000007798 UP000008792 UP000008711 UP000001070 UP000075884 UP000009192 UP000036403 UP000008820 UP000075885 UP000192221 UP000002282 UP000000304 UP000000803 UP000076407 UP000268350 UP000075920 UP000075901 UP000053097 UP000279307 UP000183832 UP000001292 UP000078540 UP000005205 UP000007801 UP000215335 UP000001819 UP000008744 UP000002320 UP000007819 UP000002358 UP000005203 UP000075809 UP000245037 UP000078541 UP000078542 UP000092460 UP000053825 UP000037069 UP000007266 UP000079169
UP000192223 UP000037510 UP000027135 UP000095301 UP000075881 UP000092444 UP000092443 UP000078200 UP000092445 UP000091820 UP000235965 UP000075880 UP000075886 UP000075883 UP000075903 UP000007062 UP000075840 UP000075882 UP000076408 UP000069272 UP000030765 UP000000311 UP000075900 UP000000673 UP000007798 UP000008792 UP000008711 UP000001070 UP000075884 UP000009192 UP000036403 UP000008820 UP000075885 UP000192221 UP000002282 UP000000304 UP000000803 UP000076407 UP000268350 UP000075920 UP000075901 UP000053097 UP000279307 UP000183832 UP000001292 UP000078540 UP000005205 UP000007801 UP000215335 UP000001819 UP000008744 UP000002320 UP000007819 UP000002358 UP000005203 UP000075809 UP000245037 UP000078541 UP000078542 UP000092460 UP000053825 UP000037069 UP000007266 UP000079169
PRIDE
Interpro
IPR038976
Ssk
ProteinModelPortal
H9JBU5
A0A2H1V6X4
A0A2A4JGU3
A0A212EUW1
A0A1D5RMU7
A0A194PI55
+ More
A8C9Z3 A0A1L8DJL8 A0A1B0GL15 E9J7N8 A0A1Q3FMB6 A0A1Q3FLV3 A0A1W4XN22 A0A1B6HMV8 A0A1B6L527 A0A1B6FGC7 A0A0L7LGW8 A0A067QYM7 T1PAC6 A0A1B6ELH2 A0A182KCR6 A0A1B0G911 A0A1A9Y043 A0A1A9UQR6 A0A1A9Z2Y7 A0A1A9WRH7 A0A2J7QHB5 A0A0C9R279 A0A182IQH7 A0A0A1XJB2 A0A1B1ZGG5 A0A182QIU4 A0A182LUW7 A0A182VLD6 Q7QDX7 A0A182HXD7 A0A182LND7 A0A182YIT5 A0A182FBV4 A0A084VUP8 T1DKF2 W8C7U8 E2ASU3 A0A182RJS8 W5JJV6 B4MKW3 A0A1Y1LLN1 B4LG14 B3NE42 B4IYP9 A0A182NDF8 B4KV85 A0A0J7K5N8 Q17HS5 A0A182PLU4 A0A1W4VU80 B4PFD8 A0APU7 C0MKE7 Q9VW87 A0A182X1N6 A0A3B0JRD9 A0A182W3X0 A0A182SUE3 A0A026X5F3 A0A1J1IGG0 B4IA52 A0A195BLG1 A0A158NLP9 A0A1S4F265 B3M4P7 A0A232F5E7 B4ITJ5 Q2M0X4 B4GQZ0 B0WA98 C4WUQ0 K7JHR0 A0A087ZXG6 A0A151WK34 A0A2P8ZPA6 A0A195FVQ8 A0A336M3J5 A0A2S2PL14 J3JYN4 A0A170YLU5 D1GZ07 D1GYQ4 A0A151IAM1 A0A1B6D7J7 A0A1B0B3K6 A0A0L7QR41 A0A1L8EHS0 A0A0L0C0F2 D6WX16 A0A0K8UXH7 A0A034WLF9 A0A3Q0IZ88 A0A146M3W1
A8C9Z3 A0A1L8DJL8 A0A1B0GL15 E9J7N8 A0A1Q3FMB6 A0A1Q3FLV3 A0A1W4XN22 A0A1B6HMV8 A0A1B6L527 A0A1B6FGC7 A0A0L7LGW8 A0A067QYM7 T1PAC6 A0A1B6ELH2 A0A182KCR6 A0A1B0G911 A0A1A9Y043 A0A1A9UQR6 A0A1A9Z2Y7 A0A1A9WRH7 A0A2J7QHB5 A0A0C9R279 A0A182IQH7 A0A0A1XJB2 A0A1B1ZGG5 A0A182QIU4 A0A182LUW7 A0A182VLD6 Q7QDX7 A0A182HXD7 A0A182LND7 A0A182YIT5 A0A182FBV4 A0A084VUP8 T1DKF2 W8C7U8 E2ASU3 A0A182RJS8 W5JJV6 B4MKW3 A0A1Y1LLN1 B4LG14 B3NE42 B4IYP9 A0A182NDF8 B4KV85 A0A0J7K5N8 Q17HS5 A0A182PLU4 A0A1W4VU80 B4PFD8 A0APU7 C0MKE7 Q9VW87 A0A182X1N6 A0A3B0JRD9 A0A182W3X0 A0A182SUE3 A0A026X5F3 A0A1J1IGG0 B4IA52 A0A195BLG1 A0A158NLP9 A0A1S4F265 B3M4P7 A0A232F5E7 B4ITJ5 Q2M0X4 B4GQZ0 B0WA98 C4WUQ0 K7JHR0 A0A087ZXG6 A0A151WK34 A0A2P8ZPA6 A0A195FVQ8 A0A336M3J5 A0A2S2PL14 J3JYN4 A0A170YLU5 D1GZ07 D1GYQ4 A0A151IAM1 A0A1B6D7J7 A0A1B0B3K6 A0A0L7QR41 A0A1L8EHS0 A0A0L0C0F2 D6WX16 A0A0K8UXH7 A0A034WLF9 A0A3Q0IZ88 A0A146M3W1
Ontologies
GO
PANTHER
Topology
Subcellular location
Apicolateral cell membrane
Specific to smooth septate junctions. With evidence from 1 publications.
Cell junction Specific to smooth septate junctions. With evidence from 1 publications.
Septate junction Specific to smooth septate junctions. With evidence from 1 publications.
Cell junction Specific to smooth septate junctions. With evidence from 1 publications.
Septate junction Specific to smooth septate junctions. With evidence from 1 publications.
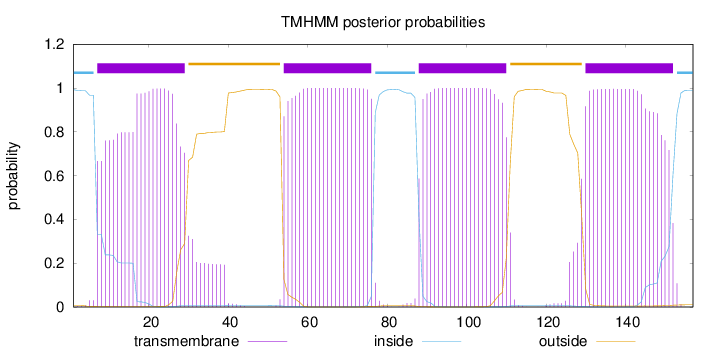
Length:
157
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
90.08884
Exp number, first 60 AAs:
28.837
Total prob of N-in:
0.99253
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 29
outside
30 - 53
TMhelix
54 - 76
inside
77 - 87
TMhelix
88 - 110
outside
111 - 129
TMhelix
130 - 152
inside
153 - 157
Population Genetic Test Statistics
Pi
251.80411
Theta
163.00117
Tajima's D
1.20989
CLR
0.864235
CSRT
0.712014399280036
Interpretation
Uncertain
