Gene
KWMTBOMO10496 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007118
Annotation
hydroxysteroid_dehydrogenase_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.767
Sequence
CDS
ATGAGTCTTGTAGCTAATACTGGAAAACTTGCGGGCCGAACGCTGTTTATTACTGGTGCTTCTCGTGGAATTGGCAAGGCTATAGCTCTGAAAGCTGCGAAAGACGGCGCAAATGTTGTAATTGCCGCGAAAACTGCCGAACCTCATCCCAAACTACCTGGCACTATTTATACCGCTGCTGAAGAAATTGAGGCTCTTGGTGGAAAGGCATTGCCATGCATTGTTGATGTTAGAGATGAGAAACAAGTGCAGAAGGCTGTTGATGAAGCCGTGAAAAAGTTCAATGGCATTGATATTCTAGTGAACAATGCATCAGCCATTTCACTAACGGGAACAGCCCAGACTGATATGAAAAGATATGATCTCATGCATAACATTAATACCAGAGGAACCTTTTTAGCGTCGAAGATATGTCTACCAGTTTTGAAAAATAGCAATCACGCCCACATTTTGAATCTGTCACCACCTCTTAACATGAACCCGTACTGGTTCTCAATTCATGTAGCATACACAATGGCCAAATACGGTATGTCTATGTGTGTGCTTGGCATGAGTGAAGAATTCAAACCATTCAATATCGGTGTCAACGCTCTGTGGCCAAAAACAGCTATAGCAACAGCGGCCATTGAAATGTTGACCGGGGACACGTCAACAAGTCGCAAGCCTGAGATCGTTTCCGACGCGGCGTACCTCATGCTCTGCAAAGACCCAAAAACGTATACCGGCAACTTTGCTATCGACGAAGACGTCGTGAAGGAAGCCGGCATCAAAGACTTATTACCCTATGCCTGTGATCCGAGTAACGTACAGAATTTGTTACCGGACTTCTTTTTGGATGTTTCCGGGGAATCCGTTGAGCCGACCATAAAGGAATCCGACACCGCAGGCGAAATCCCAGCGCTGTTTTCATTGATCGGTAAAAACCTGAGCGCCGACTTAGTGAAGAAGACTCAAGCCGTATTCCAGTTCAACGTCAAAGGTAAAGAAGAGGGAGTGTGGCATATCGACCTCAAGAACGGCGAGGGTACGTGCGGGCAGGGCGAGCCCAAGAACCCTCCGGACGCGACTCTGACGATGGACGGGAAGAACTTCACTGACATGTTCGCGGGAAAACTAAAACCGACGACAGCCTTCATGATGGGAAAACTGAAAATCAAAGGTGATCTACAAAAGGCAATGAAGCTTGAAAAGATGATGCAGTCGTTGAAGAAGAAAGCTTAG
Protein
MSLVANTGKLAGRTLFITGASRGIGKAIALKAAKDGANVVIAAKTAEPHPKLPGTIYTAAEEIEALGGKALPCIVDVRDEKQVQKAVDEAVKKFNGIDILVNNASAISLTGTAQTDMKRYDLMHNINTRGTFLASKICLPVLKNSNHAHILNLSPPLNMNPYWFSIHVAYTMAKYGMSMCVLGMSEEFKPFNIGVNALWPKTAIATAAIEMLTGDTSTSRKPEIVSDAAYLMLCKDPKTYTGNFAIDEDVVKEAGIKDLLPYACDPSNVQNLLPDFFLDVSGESVEPTIKESDTAGEIPALFSLIGKNLSADLVKKTQAVFQFNVKGKEEGVWHIDLKNGEGTCGQGEPKNPPDATLTMDGKNFTDMFAGKLKPTTAFMMGKLKIKGDLQKAMKLEKMMQSLKKKA
Summary
Uniprot
H9JC73
Q1HPV6
A0A0N1I8M8
A0A2A4JFS3
I4DMW6
A0A212FHN5
+ More
A0A2A4JH74 A0A2H1V6V2 A0A3S2M2U5 A0A194PK46 N6UB60 A0A0K8TN42 U4U808 A0A0K8UJV2 K7IYJ2 A0A0C9RT63 A0A034W7D2 A0A1B0DNZ3 A0A232ETW0 W8C9Y8 A0A2J7PSV5 A0A158NKP0 A0A1B0G7Q3 D6WW74 A0A0A1XDA3 A0A1A9V8E1 U5EZ86 A0A195DG87 D3TRX0 Q29BG6 A0A195BND5 A0A195CJ23 A0A0L0CQB6 B4NAR4 B4GNX4 A0A336MA68 A0A336M689 E2AJV5 A0A0A9WUB0 A0A1B6DJC4 A0A0L7R3K8 A0A3B0K6T9 B0WS47 A0A182J8I1 E2BLL5 A0A195FCC7 A0A3B0JYX8 A0A1S4F1N2 A0A1S4F1N5 A0A224XPT3 A0A1Q3F7J4 A0A1B6GCD4 W5J8Q7 B3LX37 Q17IA4 A0A1J1IN05 A0A2M4BP74 A0A151WRS0 Q1HQY9 A0A1I8NC44 A0A2C9GR76 Q9VB10 Q7PTS9 A0A2M4AFQ2 E9IBY2 A0A1B6HCE3 A0A0N7Z9I5 R4G5K0 A0A2M4AE67 A0A2M4BP82 A0A182KZV6 T1DNB9 A0A0M4EN11 B3P5Y8 A0A2M3Z3S5 A0A1I8PPH4 A0A2M3Z3S0 A0A2M3Z3U3 A0A182QED7 B4QY28 A0A182M4F0 B4IGU8 A0A2M4AEY5 B4PRB4 A0A182VG88 A0A182YQF8 A0A0N0BCG0 B4M622 A0A1W4V8P1 A0A1W4WE02 A0A182W363 B4JU66 A0A0C9Q8U4 A0A182XF48 A0A182U8T1 A0A182JTK9 A0A182FAD2 A0A182RI45 A0A182PLF1 E0VM98
A0A2A4JH74 A0A2H1V6V2 A0A3S2M2U5 A0A194PK46 N6UB60 A0A0K8TN42 U4U808 A0A0K8UJV2 K7IYJ2 A0A0C9RT63 A0A034W7D2 A0A1B0DNZ3 A0A232ETW0 W8C9Y8 A0A2J7PSV5 A0A158NKP0 A0A1B0G7Q3 D6WW74 A0A0A1XDA3 A0A1A9V8E1 U5EZ86 A0A195DG87 D3TRX0 Q29BG6 A0A195BND5 A0A195CJ23 A0A0L0CQB6 B4NAR4 B4GNX4 A0A336MA68 A0A336M689 E2AJV5 A0A0A9WUB0 A0A1B6DJC4 A0A0L7R3K8 A0A3B0K6T9 B0WS47 A0A182J8I1 E2BLL5 A0A195FCC7 A0A3B0JYX8 A0A1S4F1N2 A0A1S4F1N5 A0A224XPT3 A0A1Q3F7J4 A0A1B6GCD4 W5J8Q7 B3LX37 Q17IA4 A0A1J1IN05 A0A2M4BP74 A0A151WRS0 Q1HQY9 A0A1I8NC44 A0A2C9GR76 Q9VB10 Q7PTS9 A0A2M4AFQ2 E9IBY2 A0A1B6HCE3 A0A0N7Z9I5 R4G5K0 A0A2M4AE67 A0A2M4BP82 A0A182KZV6 T1DNB9 A0A0M4EN11 B3P5Y8 A0A2M3Z3S5 A0A1I8PPH4 A0A2M3Z3S0 A0A2M3Z3U3 A0A182QED7 B4QY28 A0A182M4F0 B4IGU8 A0A2M4AEY5 B4PRB4 A0A182VG88 A0A182YQF8 A0A0N0BCG0 B4M622 A0A1W4V8P1 A0A1W4WE02 A0A182W363 B4JU66 A0A0C9Q8U4 A0A182XF48 A0A182U8T1 A0A182JTK9 A0A182FAD2 A0A182RI45 A0A182PLF1 E0VM98
Pubmed
19121390
26354079
22651552
22118469
23537049
26369729
+ More
20075255 25348373 28648823 24495485 21347285 18362917 19820115 25830018 20353571 15632085 17994087 26108605 20798317 25401762 20920257 23761445 17510324 17204158 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 9087549 12364791 14747013 17210077 21282665 27129103 20966253 17550304 25244985 20566863
20075255 25348373 28648823 24495485 21347285 18362917 19820115 25830018 20353571 15632085 17994087 26108605 20798317 25401762 20920257 23761445 17510324 17204158 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 9087549 12364791 14747013 17210077 21282665 27129103 20966253 17550304 25244985 20566863
EMBL
BABH01015022
BABH01015023
DQ443296
ABF51385.1
KQ460711
KPJ12662.1
+ More
NWSH01001534 PCG70935.1 AK402634 BAM19256.1 AGBW02008482 OWR53250.1 PCG70934.1 ODYU01000980 SOQ36551.1 RSAL01000054 RVE50017.1 KQ459603 KPI93099.1 APGK01035637 KB740928 ENN77891.1 GDAI01001816 JAI15787.1 KB632064 ERL88463.1 GDHF01025689 JAI26625.1 GBYB01010691 JAG80458.1 GAKP01008333 GAKP01008332 JAC50619.1 AJVK01017924 NNAY01002209 OXU21798.1 GAMC01001744 JAC04812.1 NEVH01021922 PNF19412.1 ADTU01018942 CCAG010016890 KQ971361 EFA09196.1 GBXI01005371 JAD08921.1 GANO01001243 JAB58628.1 KQ980886 KYN11856.1 EZ424172 ADD20448.1 CM000070 EAL27032.1 KQ976439 KYM86728.1 KQ977649 KYN00725.1 JRES01000049 KNC34565.1 CH964232 EDW80878.1 CH479186 EDW38857.1 UFQT01000608 SSX25673.1 UFQT01000513 SSX24871.1 GL440100 EFN66271.1 GBHO01032598 GBRD01016710 JAG11006.1 JAG49117.1 GEDC01011497 GEDC01002023 JAS25801.1 JAS35275.1 KQ414663 KOC65336.1 OUUW01000005 SPP80711.1 DS232064 EDS33666.1 GL449036 EFN83349.1 KQ981693 KYN37857.1 SPP80710.1 GFTR01005976 JAW10450.1 GFDL01011504 JAV23541.1 GECZ01009683 JAS60086.1 ADMH02001950 ETN60346.1 CH902617 EDV43876.1 CH477241 EAT46391.1 CVRI01000054 CRL00470.1 GGFJ01005739 MBW54880.1 KQ982805 KYQ50544.1 DQ440305 ABF18338.1 EJY57419.1 APCN01005802 AE014297 AY047508 AAF56735.1 AAK77240.1 AAAB01008574 EAA03441.3 GGFK01006147 MBW39468.1 GL762137 EFZ21993.1 GECU01035424 GECU01001939 JAS72282.1 JAT05768.1 GDKW01000451 JAI56144.1 ACPB03009592 GAHY01000480 JAA77030.1 GGFK01005764 MBW39085.1 GGFJ01005736 MBW54877.1 GAMD01003020 JAA98570.1 CP012526 ALC45964.1 CH954182 EDV53388.1 GGFM01002418 MBW23169.1 GGFM01002392 MBW23143.1 GGFM01002367 MBW23118.1 AXCN02001583 CM000364 EDX14676.1 AXCM01000898 CH480837 EDW49066.1 GGFK01006025 MBW39346.1 CM000160 EDW98478.1 KQ435911 KOX68942.1 CH940652 EDW59098.1 CH916374 EDV91036.1 GBYB01010688 JAG80455.1 DS235303 EEB14504.1
NWSH01001534 PCG70935.1 AK402634 BAM19256.1 AGBW02008482 OWR53250.1 PCG70934.1 ODYU01000980 SOQ36551.1 RSAL01000054 RVE50017.1 KQ459603 KPI93099.1 APGK01035637 KB740928 ENN77891.1 GDAI01001816 JAI15787.1 KB632064 ERL88463.1 GDHF01025689 JAI26625.1 GBYB01010691 JAG80458.1 GAKP01008333 GAKP01008332 JAC50619.1 AJVK01017924 NNAY01002209 OXU21798.1 GAMC01001744 JAC04812.1 NEVH01021922 PNF19412.1 ADTU01018942 CCAG010016890 KQ971361 EFA09196.1 GBXI01005371 JAD08921.1 GANO01001243 JAB58628.1 KQ980886 KYN11856.1 EZ424172 ADD20448.1 CM000070 EAL27032.1 KQ976439 KYM86728.1 KQ977649 KYN00725.1 JRES01000049 KNC34565.1 CH964232 EDW80878.1 CH479186 EDW38857.1 UFQT01000608 SSX25673.1 UFQT01000513 SSX24871.1 GL440100 EFN66271.1 GBHO01032598 GBRD01016710 JAG11006.1 JAG49117.1 GEDC01011497 GEDC01002023 JAS25801.1 JAS35275.1 KQ414663 KOC65336.1 OUUW01000005 SPP80711.1 DS232064 EDS33666.1 GL449036 EFN83349.1 KQ981693 KYN37857.1 SPP80710.1 GFTR01005976 JAW10450.1 GFDL01011504 JAV23541.1 GECZ01009683 JAS60086.1 ADMH02001950 ETN60346.1 CH902617 EDV43876.1 CH477241 EAT46391.1 CVRI01000054 CRL00470.1 GGFJ01005739 MBW54880.1 KQ982805 KYQ50544.1 DQ440305 ABF18338.1 EJY57419.1 APCN01005802 AE014297 AY047508 AAF56735.1 AAK77240.1 AAAB01008574 EAA03441.3 GGFK01006147 MBW39468.1 GL762137 EFZ21993.1 GECU01035424 GECU01001939 JAS72282.1 JAT05768.1 GDKW01000451 JAI56144.1 ACPB03009592 GAHY01000480 JAA77030.1 GGFK01005764 MBW39085.1 GGFJ01005736 MBW54877.1 GAMD01003020 JAA98570.1 CP012526 ALC45964.1 CH954182 EDV53388.1 GGFM01002418 MBW23169.1 GGFM01002392 MBW23143.1 GGFM01002367 MBW23118.1 AXCN02001583 CM000364 EDX14676.1 AXCM01000898 CH480837 EDW49066.1 GGFK01006025 MBW39346.1 CM000160 EDW98478.1 KQ435911 KOX68942.1 CH940652 EDW59098.1 CH916374 EDV91036.1 GBYB01010688 JAG80455.1 DS235303 EEB14504.1
Proteomes
UP000005204
UP000053240
UP000218220
UP000007151
UP000283053
UP000053268
+ More
UP000019118 UP000030742 UP000002358 UP000092462 UP000215335 UP000235965 UP000005205 UP000092444 UP000007266 UP000078200 UP000078492 UP000001819 UP000078540 UP000078542 UP000037069 UP000007798 UP000008744 UP000000311 UP000053825 UP000268350 UP000002320 UP000075880 UP000008237 UP000078541 UP000000673 UP000007801 UP000008820 UP000183832 UP000075809 UP000095301 UP000075840 UP000000803 UP000007062 UP000015103 UP000075882 UP000092553 UP000008711 UP000095300 UP000075886 UP000000304 UP000075883 UP000001292 UP000002282 UP000075903 UP000076408 UP000053105 UP000008792 UP000192221 UP000192223 UP000075920 UP000001070 UP000076407 UP000075902 UP000075881 UP000069272 UP000075900 UP000075885 UP000009046
UP000019118 UP000030742 UP000002358 UP000092462 UP000215335 UP000235965 UP000005205 UP000092444 UP000007266 UP000078200 UP000078492 UP000001819 UP000078540 UP000078542 UP000037069 UP000007798 UP000008744 UP000000311 UP000053825 UP000268350 UP000002320 UP000075880 UP000008237 UP000078541 UP000000673 UP000007801 UP000008820 UP000183832 UP000075809 UP000095301 UP000075840 UP000000803 UP000007062 UP000015103 UP000075882 UP000092553 UP000008711 UP000095300 UP000075886 UP000000304 UP000075883 UP000001292 UP000002282 UP000075903 UP000076408 UP000053105 UP000008792 UP000192221 UP000192223 UP000075920 UP000001070 UP000076407 UP000075902 UP000075881 UP000069272 UP000075900 UP000075885 UP000009046
Interpro
Gene 3D
ProteinModelPortal
H9JC73
Q1HPV6
A0A0N1I8M8
A0A2A4JFS3
I4DMW6
A0A212FHN5
+ More
A0A2A4JH74 A0A2H1V6V2 A0A3S2M2U5 A0A194PK46 N6UB60 A0A0K8TN42 U4U808 A0A0K8UJV2 K7IYJ2 A0A0C9RT63 A0A034W7D2 A0A1B0DNZ3 A0A232ETW0 W8C9Y8 A0A2J7PSV5 A0A158NKP0 A0A1B0G7Q3 D6WW74 A0A0A1XDA3 A0A1A9V8E1 U5EZ86 A0A195DG87 D3TRX0 Q29BG6 A0A195BND5 A0A195CJ23 A0A0L0CQB6 B4NAR4 B4GNX4 A0A336MA68 A0A336M689 E2AJV5 A0A0A9WUB0 A0A1B6DJC4 A0A0L7R3K8 A0A3B0K6T9 B0WS47 A0A182J8I1 E2BLL5 A0A195FCC7 A0A3B0JYX8 A0A1S4F1N2 A0A1S4F1N5 A0A224XPT3 A0A1Q3F7J4 A0A1B6GCD4 W5J8Q7 B3LX37 Q17IA4 A0A1J1IN05 A0A2M4BP74 A0A151WRS0 Q1HQY9 A0A1I8NC44 A0A2C9GR76 Q9VB10 Q7PTS9 A0A2M4AFQ2 E9IBY2 A0A1B6HCE3 A0A0N7Z9I5 R4G5K0 A0A2M4AE67 A0A2M4BP82 A0A182KZV6 T1DNB9 A0A0M4EN11 B3P5Y8 A0A2M3Z3S5 A0A1I8PPH4 A0A2M3Z3S0 A0A2M3Z3U3 A0A182QED7 B4QY28 A0A182M4F0 B4IGU8 A0A2M4AEY5 B4PRB4 A0A182VG88 A0A182YQF8 A0A0N0BCG0 B4M622 A0A1W4V8P1 A0A1W4WE02 A0A182W363 B4JU66 A0A0C9Q8U4 A0A182XF48 A0A182U8T1 A0A182JTK9 A0A182FAD2 A0A182RI45 A0A182PLF1 E0VM98
A0A2A4JH74 A0A2H1V6V2 A0A3S2M2U5 A0A194PK46 N6UB60 A0A0K8TN42 U4U808 A0A0K8UJV2 K7IYJ2 A0A0C9RT63 A0A034W7D2 A0A1B0DNZ3 A0A232ETW0 W8C9Y8 A0A2J7PSV5 A0A158NKP0 A0A1B0G7Q3 D6WW74 A0A0A1XDA3 A0A1A9V8E1 U5EZ86 A0A195DG87 D3TRX0 Q29BG6 A0A195BND5 A0A195CJ23 A0A0L0CQB6 B4NAR4 B4GNX4 A0A336MA68 A0A336M689 E2AJV5 A0A0A9WUB0 A0A1B6DJC4 A0A0L7R3K8 A0A3B0K6T9 B0WS47 A0A182J8I1 E2BLL5 A0A195FCC7 A0A3B0JYX8 A0A1S4F1N2 A0A1S4F1N5 A0A224XPT3 A0A1Q3F7J4 A0A1B6GCD4 W5J8Q7 B3LX37 Q17IA4 A0A1J1IN05 A0A2M4BP74 A0A151WRS0 Q1HQY9 A0A1I8NC44 A0A2C9GR76 Q9VB10 Q7PTS9 A0A2M4AFQ2 E9IBY2 A0A1B6HCE3 A0A0N7Z9I5 R4G5K0 A0A2M4AE67 A0A2M4BP82 A0A182KZV6 T1DNB9 A0A0M4EN11 B3P5Y8 A0A2M3Z3S5 A0A1I8PPH4 A0A2M3Z3S0 A0A2M3Z3U3 A0A182QED7 B4QY28 A0A182M4F0 B4IGU8 A0A2M4AEY5 B4PRB4 A0A182VG88 A0A182YQF8 A0A0N0BCG0 B4M622 A0A1W4V8P1 A0A1W4WE02 A0A182W363 B4JU66 A0A0C9Q8U4 A0A182XF48 A0A182U8T1 A0A182JTK9 A0A182FAD2 A0A182RI45 A0A182PLF1 E0VM98
PDB
3KVO
E-value=3.74425e-94,
Score=880
Ontologies
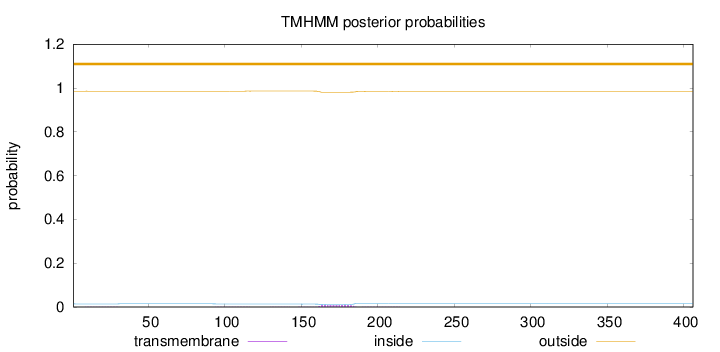
Topology
Length:
406
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.24037
Exp number, first 60 AAs:
0.00857
Total prob of N-in:
0.01428
outside
1 - 406
Population Genetic Test Statistics
Pi
351.920399
Theta
171.026777
Tajima's D
3.237244
CLR
0.692603
CSRT
0.988200589970501
Interpretation
Uncertain
