Gene
KWMTBOMO10495
Pre Gene Modal
BGIBMGA007117
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_uncharacterized_protein_LOC101745926_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.888
Sequence
CDS
ATGAAGGTGTCATGCTCCTGGGACGACGTGAAGTCTCTGCGCTCAGACATGGAGAAGCATACGTCATCGTCCATACACTTCAGACTGAAGCTACTGACGGCCGCCGCCCAGATGCAGTCCGCTCTCGGAGTGCAGGACTTAGGTCAGTTATTCCACAAGCCCCTAAGAGACTCCCACGGCACAGTGGTGCTCTCTTGCGTGAACAGCGTTAAATCTGCGAAGTCTGTGTCCGCGCTGAACTCGAGGTGGGCCCCCGTGTCCAAGCTGAGGCGGCGAGTGCTGAGCGAGGACAACACCATAGGGGAACTCCTCATGGCTTCCGTGCACGAACAGATCGCGTACCATCAGGTGTCTAGAGTGCAATTATCACGCGGGCTGTATCTCGGCTATCTGAAGCTGCAGTCCTCGGTGGAGGTGGTCCGGATCGTGGCTCCGGCGCGCGCTCCCAACGTGCCGCCTCACACCAGGGTCCGAGACAACCCGCACGTCTCAGCTGAAGAATGGGAGTACTTAAGAAATCAGTCGGGCAGATCGGATGACACCCAACAGATCCCCTCCATTAACAACTCAGAGAAGCCATCAGAATCTCAAGAGATGTTCTTCGACAACATAGCCGCTGCTGCAAGAAGACTCTTCAAAGAGATGGATATCCCTATAGACAGTGTAGCTTCCCACCGAATCTATGACTTGGAAGTCATAGAGCTGAACAATGACGTCAGCTTCGTGATGATATGCCCGCCGGCCGAACAAGCCTGCTCAGTCCCAGGACAGAAGGAGATACTGCTCCAGAAAGGCGATCTGCTCAGCTTACCGATACAAGCTTTCGAGATGATTCATCTGCAGACTTACCACACGAATATCCTCAATAAATACTCTAAGCTCAGCTGTGTCCTCGAACTGGACTTGGCCCTAGCCGCTCACTCGCACAGAGAAGCCTTCTCGTGCACGGAACTTCAGACTGCCAAAGAAAGGCTCGCTAAGTTAGAAGATCTACAGAACACTTTGAATACCGTTTGGAAGGCGGAACGGTGGTTAATGGATCTAATATGTTTCGCCAGGGACAAAGACAAGGCCGCCCAAAGCTCCGGTATATTGCTCAAGCACGTTCTAGAAAAGACTCCGACGCAAACGATTGACGGTCCCGACATAGAGACCCTGGATAGGGAGTCCAAGTTGAAGGACTTCCTTCAGATACCGTCGAGAGACGGAAAAATCACCAAAACCTCTCCGGGCCGAGGATCCTGGCCCGGGCCAGCCGGCAACGGGAATCACGCAAACTTGCACCCGGAATTCAGCAAATCCGAACAGCAACTCAGCTTGGCTCCGAGACAAACGTCCGTTTCCACGCTAAATCTGAGCAGCGCGCAGATGCTCAACGATGAAGCCATGAAATACGCCAGGAAGGGCAGCGCGGACTCCCAATACACCCAGTCTAGCACGAACTACATGAGCAGTAACTCCCAGTTCGATATCAAATCATCCGGATCTCATACTAGTATCGGCAGCAATCGCCTCCCGCCGTCCAGAAGTGAGGATACCCTGGTGCTTCAGAATGAGAACGTTGAACCGAAGCCGCGGCCTCACAGCATCAACGCGAGCTCGTCGAACTCCTCCAGCCCGCTGCTCACGGTCAAAGGCTACTACCCCGGCAGCATGATCAGCGTCAGGGCGTCCAAAGGAAACGTCTCTGAGTCCGTTCAGAGTCTATCCAGCGACTCGGAGAGCCAGAGCTGCCCCATCACGACAGTTCCTCTGAAAATCCGTCCCAAGCAATATCGTGTTCTTTCCAGCGTTATAACCTCGAAGAGTATGACTAATGTGAAGAGTTCCGATGTGGAATACACTGATACTGGTCAAGAATACCCGAAGAACAGTCTTTGCGTGAAGCGACAACCGCTTCTGGAGACACAAGAGGACGCGGAGTATAACAATAAGGTTCCTAAGACTGAGGAGACGAGGAGTGAAGTGGATGATGTGGTGAAGGGGCCTGCAGAACTGCGGACTTCGGAACAGGTAGCAGGGATACTGCAGGTGTATGCGGCCTACGAAACTGGTCTAGCGGCCGGGACGTCGCTGAAGTTACACGTGACGCCCCGCACCTCAGCCCGCGAGGTCATCGACCTGGTCGTCAAACAACTCAATATGGCCGCCGTGTCAAAAGGAAAAACTGGACCGGTTTACGGACCGGAAAAATTGAAAGACTTCTGTCTGGTGGCCGTCATAGGAGCGCGGGAGAGATGTTTGCGAGATGATTTCCGCCCGTTGCAACTTCAAAATCCGTGGAGGAAAGGACGGCTGTACGTTAGACTCAAGCATGACGTTTTGGCGGCATTGCAGCATTCGGCTAAACAGCCGGCTTACATTTAA
Protein
MKVSCSWDDVKSLRSDMEKHTSSSIHFRLKLLTAAAQMQSALGVQDLGQLFHKPLRDSHGTVVLSCVNSVKSAKSVSALNSRWAPVSKLRRRVLSEDNTIGELLMASVHEQIAYHQVSRVQLSRGLYLGYLKLQSSVEVVRIVAPARAPNVPPHTRVRDNPHVSAEEWEYLRNQSGRSDDTQQIPSINNSEKPSESQEMFFDNIAAAARRLFKEMDIPIDSVASHRIYDLEVIELNNDVSFVMICPPAEQACSVPGQKEILLQKGDLLSLPIQAFEMIHLQTYHTNILNKYSKLSCVLELDLALAAHSHREAFSCTELQTAKERLAKLEDLQNTLNTVWKAERWLMDLICFARDKDKAAQSSGILLKHVLEKTPTQTIDGPDIETLDRESKLKDFLQIPSRDGKITKTSPGRGSWPGPAGNGNHANLHPEFSKSEQQLSLAPRQTSVSTLNLSSAQMLNDEAMKYARKGSADSQYTQSSTNYMSSNSQFDIKSSGSHTSIGSNRLPPSRSEDTLVLQNENVEPKPRPHSINASSSNSSSPLLTVKGYYPGSMISVRASKGNVSESVQSLSSDSESQSCPITTVPLKIRPKQYRVLSSVITSKSMTNVKSSDVEYTDTGQEYPKNSLCVKRQPLLETQEDAEYNNKVPKTEETRSEVDDVVKGPAELRTSEQVAGILQVYAAYETGLAAGTSLKLHVTPRTSAREVIDLVVKQLNMAAVSKGKTGPVYGPEKLKDFCLVAVIGARERCLRDDFRPLQLQNPWRKGRLYVRLKHDVLAALQHSAKQPAYI
Summary
Uniprot
H9JC72
A0A2A4JFT7
A0A194PJ82
A0A2H1X3D2
A0A0L7L3C2
A0A0N0PCA7
+ More
A0A139WCN1 A0A139WCK7 A0A1B0CQA0 A0A1W4WMM6 A0A1Y1JT66 A0A1Y1JW94 A0A1W4WNN0 A0A1Y1JT83 A0A1W4WCK8 A0A026W0G9 E2AWX2 A0A154PPR6 A0A151IG01 A0A151I630 E2BAM3 A0A088AR62 A0A158P194 A0A2A3EAM1 A0A151JXB9 A0A0L7REU5 A0A151IRH6 A0A0J7KYN4 A0A1Y1JZN7 A0A1Y1K138 B0WVL8 A0A1Y1JWD3 A0A182MSW6 A0A0N0U4E2 U4UKX8 A0A0P8XYQ0 A0A0P9AQ19 B3LUZ7 A0A0P8XYP6 A0A0B4K6W6 Q9VCW7 X4YX01 A0A0B4LHH4 A0A0B4K6D0 A0A0R1E4E8 B4PMD2 A0A0R1EAH9 A0A0R1E4C2 D0Z761 B3P8L7 A0A0R1E9B7 B4R0N8 A0A3B0KRA5 A0A0Q9XFX6 N6T6X4 B4KCY5 A0A0Q9XDV6 A0A0Q9XCP0 A0A0R3NMA5 Q299F6 A0A0R3NMA3 B4G543 A0A0R3NGN8 I5ANS3 A0A1W4VKZ6 A0A1W4V8L5 A0A1I8PR08 A0A1W4V870 A0A1I8PR46 A0A0M4EKM5 A0A3L8DLX6 B4M645 A0A0Q9WEE6 A0A0T6BE22 A0A1Y1JWE2 A0A182GZZ3 A0A151WND1 A0A1Y1JYH7 A0A1Y1JT68 A0A0C9RB68 Q16LK0 A0A0C9QTV4 A0A0K8WK23 A0A0C9R7V9 F4W8S9 A0A0A1WPL2 W8BR22 A0A034VFB3 A0A0K8VFN4 W8B4H6
A0A139WCN1 A0A139WCK7 A0A1B0CQA0 A0A1W4WMM6 A0A1Y1JT66 A0A1Y1JW94 A0A1W4WNN0 A0A1Y1JT83 A0A1W4WCK8 A0A026W0G9 E2AWX2 A0A154PPR6 A0A151IG01 A0A151I630 E2BAM3 A0A088AR62 A0A158P194 A0A2A3EAM1 A0A151JXB9 A0A0L7REU5 A0A151IRH6 A0A0J7KYN4 A0A1Y1JZN7 A0A1Y1K138 B0WVL8 A0A1Y1JWD3 A0A182MSW6 A0A0N0U4E2 U4UKX8 A0A0P8XYQ0 A0A0P9AQ19 B3LUZ7 A0A0P8XYP6 A0A0B4K6W6 Q9VCW7 X4YX01 A0A0B4LHH4 A0A0B4K6D0 A0A0R1E4E8 B4PMD2 A0A0R1EAH9 A0A0R1E4C2 D0Z761 B3P8L7 A0A0R1E9B7 B4R0N8 A0A3B0KRA5 A0A0Q9XFX6 N6T6X4 B4KCY5 A0A0Q9XDV6 A0A0Q9XCP0 A0A0R3NMA5 Q299F6 A0A0R3NMA3 B4G543 A0A0R3NGN8 I5ANS3 A0A1W4VKZ6 A0A1W4V8L5 A0A1I8PR08 A0A1W4V870 A0A1I8PR46 A0A0M4EKM5 A0A3L8DLX6 B4M645 A0A0Q9WEE6 A0A0T6BE22 A0A1Y1JWE2 A0A182GZZ3 A0A151WND1 A0A1Y1JYH7 A0A1Y1JT68 A0A0C9RB68 Q16LK0 A0A0C9QTV4 A0A0K8WK23 A0A0C9R7V9 F4W8S9 A0A0A1WPL2 W8BR22 A0A034VFB3 A0A0K8VFN4 W8B4H6
Pubmed
EMBL
BABH01015020
BABH01015021
NWSH01001534
PCG70937.1
KQ459603
KPI93098.1
+ More
ODYU01013151 SOQ59843.1 JTDY01003201 KOB69962.1 KQ460711 KPJ12661.1 KQ971363 KYB25680.1 KYB25679.1 AJWK01023294 AJWK01023295 GEZM01101593 JAV52474.1 GEZM01101589 JAV52478.1 GEZM01101585 JAV52482.1 KK107503 EZA49562.1 GL443425 EFN62092.1 KQ435010 KZC13737.1 KQ977743 KYN00199.1 KQ976402 KYM92302.1 GL446793 EFN87249.1 ADTU01006125 ADTU01006126 ADTU01006127 ADTU01006128 ADTU01006129 ADTU01006130 ADTU01006131 ADTU01006132 ADTU01006133 ADTU01006134 KZ288313 PBC28342.1 KQ981619 KYN39237.1 KQ414608 KOC69350.1 KQ981127 KYN09296.1 LBMM01002004 KMQ95408.1 GEZM01101591 JAV52476.1 GEZM01101590 JAV52477.1 DS232128 EDS35625.1 GEZM01101596 JAV52471.1 AXCM01000025 AXCM01000026 KQ435837 KOX71478.1 KB632255 ERL90660.1 CH902617 KPU79745.1 KPU79744.1 KPU79746.1 EDV42469.2 KPU79747.1 AE014297 AFH06561.1 AAF56038.2 ADV37357.1 KJ131166 AHN57468.1 AHV83438.1 AHN57469.1 AFH06562.1 AFH06563.1 CM000160 KRK04088.1 KRK04089.1 EDW98037.2 KRK04087.1 KRK04086.1 KRK04090.1 BT100308 ACZ52620.1 CH954182 EDV54112.1 KRK04084.1 KRK04085.1 CM000364 EDX13961.1 OUUW01000013 SPP87771.1 CH933806 KRG02451.1 APGK01041587 APGK01041588 APGK01041589 APGK01041590 APGK01041591 APGK01041592 APGK01041593 KB740994 ENN75979.1 EDW17007.2 KRG02449.1 KRG02450.1 CM000070 KRT00257.1 KRT00259.1 EAL27747.3 KRT00256.1 CH479179 EDW24709.1 KRT00258.1 EIM52608.2 CP012526 ALC47784.1 QOIP01000006 RLU21440.1 CH940652 EDW59121.2 KRF78703.1 KRF78704.1 KRF78705.1 LJIG01001435 KRT85505.1 GEZM01101594 GEZM01101586 JAV52481.1 JXUM01022082 JXUM01022083 KQ560620 KXJ81581.1 KQ982916 KYQ49301.1 GEZM01101592 GEZM01101587 JAV52475.1 GEZM01101595 GEZM01101588 JAV52479.1 GBYB01004116 JAG73883.1 CH477903 EAT35190.1 GBYB01004117 JAG73884.1 GDHF01000877 JAI51437.1 GBYB01004115 JAG73882.1 GL887917 EGI69396.1 GBXI01013964 JAD00328.1 GAMC01014611 JAB91944.1 GAKP01018170 JAC40782.1 GDHF01014974 JAI37340.1 GAMC01014612 JAB91943.1
ODYU01013151 SOQ59843.1 JTDY01003201 KOB69962.1 KQ460711 KPJ12661.1 KQ971363 KYB25680.1 KYB25679.1 AJWK01023294 AJWK01023295 GEZM01101593 JAV52474.1 GEZM01101589 JAV52478.1 GEZM01101585 JAV52482.1 KK107503 EZA49562.1 GL443425 EFN62092.1 KQ435010 KZC13737.1 KQ977743 KYN00199.1 KQ976402 KYM92302.1 GL446793 EFN87249.1 ADTU01006125 ADTU01006126 ADTU01006127 ADTU01006128 ADTU01006129 ADTU01006130 ADTU01006131 ADTU01006132 ADTU01006133 ADTU01006134 KZ288313 PBC28342.1 KQ981619 KYN39237.1 KQ414608 KOC69350.1 KQ981127 KYN09296.1 LBMM01002004 KMQ95408.1 GEZM01101591 JAV52476.1 GEZM01101590 JAV52477.1 DS232128 EDS35625.1 GEZM01101596 JAV52471.1 AXCM01000025 AXCM01000026 KQ435837 KOX71478.1 KB632255 ERL90660.1 CH902617 KPU79745.1 KPU79744.1 KPU79746.1 EDV42469.2 KPU79747.1 AE014297 AFH06561.1 AAF56038.2 ADV37357.1 KJ131166 AHN57468.1 AHV83438.1 AHN57469.1 AFH06562.1 AFH06563.1 CM000160 KRK04088.1 KRK04089.1 EDW98037.2 KRK04087.1 KRK04086.1 KRK04090.1 BT100308 ACZ52620.1 CH954182 EDV54112.1 KRK04084.1 KRK04085.1 CM000364 EDX13961.1 OUUW01000013 SPP87771.1 CH933806 KRG02451.1 APGK01041587 APGK01041588 APGK01041589 APGK01041590 APGK01041591 APGK01041592 APGK01041593 KB740994 ENN75979.1 EDW17007.2 KRG02449.1 KRG02450.1 CM000070 KRT00257.1 KRT00259.1 EAL27747.3 KRT00256.1 CH479179 EDW24709.1 KRT00258.1 EIM52608.2 CP012526 ALC47784.1 QOIP01000006 RLU21440.1 CH940652 EDW59121.2 KRF78703.1 KRF78704.1 KRF78705.1 LJIG01001435 KRT85505.1 GEZM01101594 GEZM01101586 JAV52481.1 JXUM01022082 JXUM01022083 KQ560620 KXJ81581.1 KQ982916 KYQ49301.1 GEZM01101592 GEZM01101587 JAV52475.1 GEZM01101595 GEZM01101588 JAV52479.1 GBYB01004116 JAG73883.1 CH477903 EAT35190.1 GBYB01004117 JAG73884.1 GDHF01000877 JAI51437.1 GBYB01004115 JAG73882.1 GL887917 EGI69396.1 GBXI01013964 JAD00328.1 GAMC01014611 JAB91944.1 GAKP01018170 JAC40782.1 GDHF01014974 JAI37340.1 GAMC01014612 JAB91943.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000037510
UP000053240
UP000007266
+ More
UP000092461 UP000192223 UP000053097 UP000000311 UP000076502 UP000078542 UP000078540 UP000008237 UP000005203 UP000005205 UP000242457 UP000078541 UP000053825 UP000078492 UP000036403 UP000002320 UP000075883 UP000053105 UP000030742 UP000007801 UP000000803 UP000002282 UP000008711 UP000000304 UP000268350 UP000009192 UP000019118 UP000001819 UP000008744 UP000192221 UP000095300 UP000092553 UP000279307 UP000008792 UP000069940 UP000249989 UP000075809 UP000008820 UP000007755
UP000092461 UP000192223 UP000053097 UP000000311 UP000076502 UP000078542 UP000078540 UP000008237 UP000005203 UP000005205 UP000242457 UP000078541 UP000053825 UP000078492 UP000036403 UP000002320 UP000075883 UP000053105 UP000030742 UP000007801 UP000000803 UP000002282 UP000008711 UP000000304 UP000268350 UP000009192 UP000019118 UP000001819 UP000008744 UP000192221 UP000095300 UP000092553 UP000279307 UP000008792 UP000069940 UP000249989 UP000075809 UP000008820 UP000007755
Interpro
Gene 3D
ProteinModelPortal
H9JC72
A0A2A4JFT7
A0A194PJ82
A0A2H1X3D2
A0A0L7L3C2
A0A0N0PCA7
+ More
A0A139WCN1 A0A139WCK7 A0A1B0CQA0 A0A1W4WMM6 A0A1Y1JT66 A0A1Y1JW94 A0A1W4WNN0 A0A1Y1JT83 A0A1W4WCK8 A0A026W0G9 E2AWX2 A0A154PPR6 A0A151IG01 A0A151I630 E2BAM3 A0A088AR62 A0A158P194 A0A2A3EAM1 A0A151JXB9 A0A0L7REU5 A0A151IRH6 A0A0J7KYN4 A0A1Y1JZN7 A0A1Y1K138 B0WVL8 A0A1Y1JWD3 A0A182MSW6 A0A0N0U4E2 U4UKX8 A0A0P8XYQ0 A0A0P9AQ19 B3LUZ7 A0A0P8XYP6 A0A0B4K6W6 Q9VCW7 X4YX01 A0A0B4LHH4 A0A0B4K6D0 A0A0R1E4E8 B4PMD2 A0A0R1EAH9 A0A0R1E4C2 D0Z761 B3P8L7 A0A0R1E9B7 B4R0N8 A0A3B0KRA5 A0A0Q9XFX6 N6T6X4 B4KCY5 A0A0Q9XDV6 A0A0Q9XCP0 A0A0R3NMA5 Q299F6 A0A0R3NMA3 B4G543 A0A0R3NGN8 I5ANS3 A0A1W4VKZ6 A0A1W4V8L5 A0A1I8PR08 A0A1W4V870 A0A1I8PR46 A0A0M4EKM5 A0A3L8DLX6 B4M645 A0A0Q9WEE6 A0A0T6BE22 A0A1Y1JWE2 A0A182GZZ3 A0A151WND1 A0A1Y1JYH7 A0A1Y1JT68 A0A0C9RB68 Q16LK0 A0A0C9QTV4 A0A0K8WK23 A0A0C9R7V9 F4W8S9 A0A0A1WPL2 W8BR22 A0A034VFB3 A0A0K8VFN4 W8B4H6
A0A139WCN1 A0A139WCK7 A0A1B0CQA0 A0A1W4WMM6 A0A1Y1JT66 A0A1Y1JW94 A0A1W4WNN0 A0A1Y1JT83 A0A1W4WCK8 A0A026W0G9 E2AWX2 A0A154PPR6 A0A151IG01 A0A151I630 E2BAM3 A0A088AR62 A0A158P194 A0A2A3EAM1 A0A151JXB9 A0A0L7REU5 A0A151IRH6 A0A0J7KYN4 A0A1Y1JZN7 A0A1Y1K138 B0WVL8 A0A1Y1JWD3 A0A182MSW6 A0A0N0U4E2 U4UKX8 A0A0P8XYQ0 A0A0P9AQ19 B3LUZ7 A0A0P8XYP6 A0A0B4K6W6 Q9VCW7 X4YX01 A0A0B4LHH4 A0A0B4K6D0 A0A0R1E4E8 B4PMD2 A0A0R1EAH9 A0A0R1E4C2 D0Z761 B3P8L7 A0A0R1E9B7 B4R0N8 A0A3B0KRA5 A0A0Q9XFX6 N6T6X4 B4KCY5 A0A0Q9XDV6 A0A0Q9XCP0 A0A0R3NMA5 Q299F6 A0A0R3NMA3 B4G543 A0A0R3NGN8 I5ANS3 A0A1W4VKZ6 A0A1W4V8L5 A0A1I8PR08 A0A1W4V870 A0A1I8PR46 A0A0M4EKM5 A0A3L8DLX6 B4M645 A0A0Q9WEE6 A0A0T6BE22 A0A1Y1JWE2 A0A182GZZ3 A0A151WND1 A0A1Y1JYH7 A0A1Y1JT68 A0A0C9RB68 Q16LK0 A0A0C9QTV4 A0A0K8WK23 A0A0C9R7V9 F4W8S9 A0A0A1WPL2 W8BR22 A0A034VFB3 A0A0K8VFN4 W8B4H6
Ontologies
GO
PANTHER
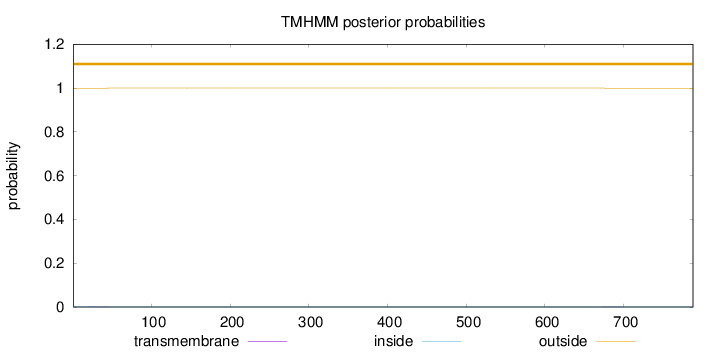
Topology
Length:
788
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02043
Exp number, first 60 AAs:
0.01295
Total prob of N-in:
0.00085
outside
1 - 788
Population Genetic Test Statistics
Pi
253.4665
Theta
166.887427
Tajima's D
1.126086
CLR
0.22807
CSRT
0.693265336733163
Interpretation
Uncertain
