Gene
KWMTBOMO10492 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006993
Annotation
PREDICTED:_multiple_inositol_polyphosphate_phosphatase_1-like_isoform_X2_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.752
Sequence
CDS
ATGCGGTGTGCCTTCATCCTGCTGATGGCAGCTTTGTGCGCTCGAGCCGACGAGATCTGCCTTACGTCTGAAGAGGACCCATATTTGCTATTCGGAACGAAGACCGCGTACATCTTCTCCAATCGTGGCATCCCAAACGCTAAGGCACATGAAGTACCAGGATGCCAACCCGTAGCCTTTTGGCTCCTCAGTCGTCACGGCTCGCATAACCCTGAAGCTAACGAGACTGCTGAACTCCAACAACTGGTCGATCTAAAGAACAACATCGTCACGAACTACAAGAATGGAAACTTCAGGAACACTAATCATCGAATCTGCACGGCCGACTTTAACTTGCTGGAACGTTGGAACTGGAAACAGAACAGTAGCACGGCCGGCGATCTCACCAGCGACGGATACGTGACGACGCAGCAGTTGGCACAGGCCTGGAAGCAGCGGTTCCCTGGACTATTGACGCCGAACACTTACAACTATTTGTTCAAATTCGTCGAAGAACCACGGTCTAGCAACTCGTTTAGAGCGTTCTCGGAAAGTTTATTCGGAGGCCAGAACGAAGGTTTCGATGCCCCCAAAGAGAACGACGAAAAACTGCTAAGGCCACTCAAGTACTGCGACGCTTGGAAGAACCAAGTCGACGAGAACAACAAGACTTTAGAACAGAAAAACATATTTGAATCGAAGCAGGAATATAAAGAGATGATTTCGAACATTTCCCTGAGACTGGGCTTCAACTACGACCTTCAGAAGAACACAGTGTTGAGCATCTACCAGATGTGCCGATACAACAAGGCGTGGGACGTGACGCAGATCTCGCCATGGTGCGCTGCGTTCACGCGTGAAGACTTAAAACGCTTGGAGTACGCCGAGGATTTGGAAACTTATTACAAATACGGTTACGGATCCACAGTCAGTGAGAAAATCGGCTGTAGCATTATTAAGGATATGGTTGACTTCTTCAAGAACCACGCTGAACGAGAGTTACGTCAACAACCAAAAGCCATGATCCAACTGACGGACTACCCGATGATCCTGATGGCTCTGACCAAGCTCGGGTCGCGGCAGGACTCGGCCCCCATCACGGGAGACAACTACCACACAAACACCGCGCAGGCCCGCAAGTGGAGCTCCTCCAACATGGTCCCATTTAATGCTAACATAGCTGCTGTTTTGTACAAGTGA
Protein
MRCAFILLMAALCARADEICLTSEEDPYLLFGTKTAYIFSNRGIPNAKAHEVPGCQPVAFWLLSRHGSHNPEANETAELQQLVDLKNNIVTNYKNGNFRNTNHRICTADFNLLERWNWKQNSSTAGDLTSDGYVTTQQLAQAWKQRFPGLLTPNTYNYLFKFVEEPRSSNSFRAFSESLFGGQNEGFDAPKENDEKLLRPLKYCDAWKNQVDENNKTLEQKNIFESKQEYKEMISNISLRLGFNYDLQKNTVLSIYQMCRYNKAWDVTQISPWCAAFTREDLKRLEYAEDLETYYKYGYGSTVSEKIGCSIIKDMVDFFKNHAERELRQQPKAMIQLTDYPMILMALTKLGSRQDSAPITGDNYHTNTAQARKWSSSNMVPFNANIAAVLYK
Summary
Similarity
Belongs to the histidine acid phosphatase family.
Uniprot
H9JBU8
A0A2H1VIP3
A0A194PK41
A0A3S2NI31
A0A194QTQ3
A0A2J7RDK1
+ More
A0A2J7RDJ9 A0A067R8V8 A0A2P8XRY2 A0A2J7RDL5 A0A2J7RDL3 A0A067R6V0 D6WUK9 A0A2J7RDK4 A0A1L8DZQ9 A0A1L8DZV2 E0VT88 A0A158NM28 U4UGG7 N6UIY3 A0A067RID8 F4WHR4 U5EX79 A0A151XHW5 A0A195B4H3 A0A195CX09 A0A154P0U4 A0A151IXB5 A0A088AJX2 A0A151JXQ6 E2B471 A0A2J7RDK5 A0A1B6MC10 A0A1B6KA08 A0A2J7RDK9 E1ZUZ6 E9G9W2 A0A2J7RDK2 A0A1B6JPG7 A0A1A9XB51 A0A310SIY4 A0A0P4YQH5 A0A1B0B6P5 A0A026WG54 A0A1B6DXK1 A0A1B6GTD8 A0A1B6F1J4 A0A1B6GAP1 A0A1B6DSF1 A0A1B6GW26 A0A0L7QR89 A0A1B6IKU5 E9IIG1 A0A0P6BS44 A0A2J7RDK7 D3TLX8 A0A0P6ITU3 A0A1A9URP2 A0A1A9WLE4 Q16FA7 A0A023ESJ6 Q16ES7 A0A023EVE1 A0A182H850 A0A1B6EIW9 A0A1B0CIG3 B3NHY8 A0A023ET15 B4HJP3 A0A336LK28 B4PKD5 B4QMU7 B0WX08 A0A232FCC4 A0A2S2Q2M4 T1IET8 A0A1Q3FIT4 A0A2P8YWZ2 B4MMG9 Q16PV9 A0A1S4FTF5 A0A2M4BLH7 A0A1W4UGU4 A0A2M4BMC2 A0A3B0KFI0 A0A0M4EDB8 A0A2M4AIN7 A0A2M4AJ04 A0A2M4AIX1 A0A0M4E967 A0A0M4E9Y7 A0A1B0DD06 B3M4H3 A0A0L0C1T2 B4H209 A0A182FDV7
A0A2J7RDJ9 A0A067R8V8 A0A2P8XRY2 A0A2J7RDL5 A0A2J7RDL3 A0A067R6V0 D6WUK9 A0A2J7RDK4 A0A1L8DZQ9 A0A1L8DZV2 E0VT88 A0A158NM28 U4UGG7 N6UIY3 A0A067RID8 F4WHR4 U5EX79 A0A151XHW5 A0A195B4H3 A0A195CX09 A0A154P0U4 A0A151IXB5 A0A088AJX2 A0A151JXQ6 E2B471 A0A2J7RDK5 A0A1B6MC10 A0A1B6KA08 A0A2J7RDK9 E1ZUZ6 E9G9W2 A0A2J7RDK2 A0A1B6JPG7 A0A1A9XB51 A0A310SIY4 A0A0P4YQH5 A0A1B0B6P5 A0A026WG54 A0A1B6DXK1 A0A1B6GTD8 A0A1B6F1J4 A0A1B6GAP1 A0A1B6DSF1 A0A1B6GW26 A0A0L7QR89 A0A1B6IKU5 E9IIG1 A0A0P6BS44 A0A2J7RDK7 D3TLX8 A0A0P6ITU3 A0A1A9URP2 A0A1A9WLE4 Q16FA7 A0A023ESJ6 Q16ES7 A0A023EVE1 A0A182H850 A0A1B6EIW9 A0A1B0CIG3 B3NHY8 A0A023ET15 B4HJP3 A0A336LK28 B4PKD5 B4QMU7 B0WX08 A0A232FCC4 A0A2S2Q2M4 T1IET8 A0A1Q3FIT4 A0A2P8YWZ2 B4MMG9 Q16PV9 A0A1S4FTF5 A0A2M4BLH7 A0A1W4UGU4 A0A2M4BMC2 A0A3B0KFI0 A0A0M4EDB8 A0A2M4AIN7 A0A2M4AJ04 A0A2M4AIX1 A0A0M4E967 A0A0M4E9Y7 A0A1B0DD06 B3M4H3 A0A0L0C1T2 B4H209 A0A182FDV7
Pubmed
EMBL
BABH01014975
BABH01014976
ODYU01002780
SOQ40697.1
KQ459603
KPI93094.1
+ More
RSAL01000031 RVE51609.1 KQ461137 KPJ08847.1 NEVH01005288 PNF38916.1 PNF38917.1 KK852663 KDR19043.1 PYGN01001450 PSN34766.1 PNF38918.1 PNF38919.1 KDR19047.1 KQ971357 EFA08487.2 PNF38911.1 GFDF01002157 JAV11927.1 GFDF01002158 JAV11926.1 DS235760 EEB16594.1 ADTU01020207 ADTU01020208 KB632286 ERL91433.1 APGK01018215 KB740071 ENN81710.1 KDR19044.1 GL888167 EGI66271.1 GANO01002665 JAB57206.1 KQ982109 KYQ59996.1 KQ976604 KYM79398.1 KQ977171 KYN05185.1 KQ434796 KZC05546.1 KQ980813 KYN12703.1 KQ981561 KYN39831.1 GL445515 EFN89488.1 PNF38908.1 GEBQ01006511 JAT33466.1 GEBQ01031695 JAT08282.1 PNF38912.1 GL434335 EFN74994.1 GL732536 EFX83810.1 PNF38913.1 GECU01027339 GECU01006669 JAS80367.1 JAT01038.1 KQ759847 OAD62581.1 GDIP01224285 GDIP01124790 JAI99116.1 JXJN01009180 KK107238 EZA54626.1 GEDC01006884 JAS30414.1 GECZ01015696 GECZ01004059 JAS54073.1 JAS65710.1 GECZ01025679 JAS44090.1 GECZ01010277 JAS59492.1 GEDC01008715 JAS28583.1 GECZ01003135 JAS66634.1 KQ414784 KOC61016.1 GECU01020168 JAS87538.1 GL763491 EFZ19634.1 GDIP01010892 JAM92823.1 PNF38923.1 CCAG010004511 EZ422430 ADD18706.1 GDIQ01017791 JAN76946.1 CH478474 EAT32917.1 GAPW01001348 JAC12250.1 CH478594 EAT32739.1 GAPW01001159 JAC12439.1 JXUM01117635 JXUM01117636 GECZ01031900 JAS37869.1 AJWK01013310 AJWK01013311 AJWK01013312 AJWK01013313 CH954178 EDV52074.1 GAPW01001158 JAC12440.1 CH480815 EDW41770.1 UFQS01000040 UFQT01000040 SSW98168.1 SSX18554.1 CM000159 EDW94833.1 CM000363 CM002912 EDX10739.1 KMZ00065.1 DS232155 EDS36269.1 NNAY01000444 OXU28335.1 GGMS01002678 MBY71881.1 ACPB03015834 GFDL01007566 JAV27479.1 PYGN01000310 PSN48762.1 CH963847 EDW73314.1 CH477771 EAT36389.1 GGFJ01004776 MBW53917.1 GGFJ01004777 MBW53918.1 OUUW01000012 SPP87110.1 CP012525 ALC43571.1 GGFK01007323 MBW40644.1 GGFK01007443 MBW40764.1 GGFK01007418 MBW40739.1 ALC43578.1 ALC43991.1 AJVK01005308 CH902618 EDV40467.1 JRES01000997 KNC26265.1 CH479203 EDW30361.1
RSAL01000031 RVE51609.1 KQ461137 KPJ08847.1 NEVH01005288 PNF38916.1 PNF38917.1 KK852663 KDR19043.1 PYGN01001450 PSN34766.1 PNF38918.1 PNF38919.1 KDR19047.1 KQ971357 EFA08487.2 PNF38911.1 GFDF01002157 JAV11927.1 GFDF01002158 JAV11926.1 DS235760 EEB16594.1 ADTU01020207 ADTU01020208 KB632286 ERL91433.1 APGK01018215 KB740071 ENN81710.1 KDR19044.1 GL888167 EGI66271.1 GANO01002665 JAB57206.1 KQ982109 KYQ59996.1 KQ976604 KYM79398.1 KQ977171 KYN05185.1 KQ434796 KZC05546.1 KQ980813 KYN12703.1 KQ981561 KYN39831.1 GL445515 EFN89488.1 PNF38908.1 GEBQ01006511 JAT33466.1 GEBQ01031695 JAT08282.1 PNF38912.1 GL434335 EFN74994.1 GL732536 EFX83810.1 PNF38913.1 GECU01027339 GECU01006669 JAS80367.1 JAT01038.1 KQ759847 OAD62581.1 GDIP01224285 GDIP01124790 JAI99116.1 JXJN01009180 KK107238 EZA54626.1 GEDC01006884 JAS30414.1 GECZ01015696 GECZ01004059 JAS54073.1 JAS65710.1 GECZ01025679 JAS44090.1 GECZ01010277 JAS59492.1 GEDC01008715 JAS28583.1 GECZ01003135 JAS66634.1 KQ414784 KOC61016.1 GECU01020168 JAS87538.1 GL763491 EFZ19634.1 GDIP01010892 JAM92823.1 PNF38923.1 CCAG010004511 EZ422430 ADD18706.1 GDIQ01017791 JAN76946.1 CH478474 EAT32917.1 GAPW01001348 JAC12250.1 CH478594 EAT32739.1 GAPW01001159 JAC12439.1 JXUM01117635 JXUM01117636 GECZ01031900 JAS37869.1 AJWK01013310 AJWK01013311 AJWK01013312 AJWK01013313 CH954178 EDV52074.1 GAPW01001158 JAC12440.1 CH480815 EDW41770.1 UFQS01000040 UFQT01000040 SSW98168.1 SSX18554.1 CM000159 EDW94833.1 CM000363 CM002912 EDX10739.1 KMZ00065.1 DS232155 EDS36269.1 NNAY01000444 OXU28335.1 GGMS01002678 MBY71881.1 ACPB03015834 GFDL01007566 JAV27479.1 PYGN01000310 PSN48762.1 CH963847 EDW73314.1 CH477771 EAT36389.1 GGFJ01004776 MBW53917.1 GGFJ01004777 MBW53918.1 OUUW01000012 SPP87110.1 CP012525 ALC43571.1 GGFK01007323 MBW40644.1 GGFK01007443 MBW40764.1 GGFK01007418 MBW40739.1 ALC43578.1 ALC43991.1 AJVK01005308 CH902618 EDV40467.1 JRES01000997 KNC26265.1 CH479203 EDW30361.1
Proteomes
UP000005204
UP000053268
UP000283053
UP000053240
UP000235965
UP000027135
+ More
UP000245037 UP000007266 UP000009046 UP000005205 UP000030742 UP000019118 UP000007755 UP000075809 UP000078540 UP000078542 UP000076502 UP000078492 UP000005203 UP000078541 UP000008237 UP000000311 UP000000305 UP000092443 UP000092460 UP000053097 UP000053825 UP000092444 UP000078200 UP000091820 UP000008820 UP000069940 UP000092461 UP000008711 UP000001292 UP000002282 UP000000304 UP000002320 UP000215335 UP000015103 UP000007798 UP000192221 UP000268350 UP000092553 UP000092462 UP000007801 UP000037069 UP000008744 UP000069272
UP000245037 UP000007266 UP000009046 UP000005205 UP000030742 UP000019118 UP000007755 UP000075809 UP000078540 UP000078542 UP000076502 UP000078492 UP000005203 UP000078541 UP000008237 UP000000311 UP000000305 UP000092443 UP000092460 UP000053097 UP000053825 UP000092444 UP000078200 UP000091820 UP000008820 UP000069940 UP000092461 UP000008711 UP000001292 UP000002282 UP000000304 UP000002320 UP000215335 UP000015103 UP000007798 UP000192221 UP000268350 UP000092553 UP000092462 UP000007801 UP000037069 UP000008744 UP000069272
PRIDE
Pfam
Interpro
IPR000560
His_Pase_clade-2
+ More
IPR016274 Histidine_acid_Pase_euk
IPR029033 His_PPase_superfam
IPR033379 Acid_Pase_AS
IPR037278 ARFGAP/RecO
IPR038508 ArfGAP_dom_sf
IPR001164 ArfGAP_dom
IPR005334 Tctex-1-like
IPR038586 Tctex-1-like_sf
IPR001611 Leu-rich_rpt
IPR003591 Leu-rich_rpt_typical-subtyp
IPR000372 LRRNT
IPR032675 LRR_dom_sf
IPR020568 Ribosomal_S5_D2-typ_fold
IPR001404 Hsp90_fam
IPR037196 HSP90_C
IPR016274 Histidine_acid_Pase_euk
IPR029033 His_PPase_superfam
IPR033379 Acid_Pase_AS
IPR037278 ARFGAP/RecO
IPR038508 ArfGAP_dom_sf
IPR001164 ArfGAP_dom
IPR005334 Tctex-1-like
IPR038586 Tctex-1-like_sf
IPR001611 Leu-rich_rpt
IPR003591 Leu-rich_rpt_typical-subtyp
IPR000372 LRRNT
IPR032675 LRR_dom_sf
IPR020568 Ribosomal_S5_D2-typ_fold
IPR001404 Hsp90_fam
IPR037196 HSP90_C
ProteinModelPortal
H9JBU8
A0A2H1VIP3
A0A194PK41
A0A3S2NI31
A0A194QTQ3
A0A2J7RDK1
+ More
A0A2J7RDJ9 A0A067R8V8 A0A2P8XRY2 A0A2J7RDL5 A0A2J7RDL3 A0A067R6V0 D6WUK9 A0A2J7RDK4 A0A1L8DZQ9 A0A1L8DZV2 E0VT88 A0A158NM28 U4UGG7 N6UIY3 A0A067RID8 F4WHR4 U5EX79 A0A151XHW5 A0A195B4H3 A0A195CX09 A0A154P0U4 A0A151IXB5 A0A088AJX2 A0A151JXQ6 E2B471 A0A2J7RDK5 A0A1B6MC10 A0A1B6KA08 A0A2J7RDK9 E1ZUZ6 E9G9W2 A0A2J7RDK2 A0A1B6JPG7 A0A1A9XB51 A0A310SIY4 A0A0P4YQH5 A0A1B0B6P5 A0A026WG54 A0A1B6DXK1 A0A1B6GTD8 A0A1B6F1J4 A0A1B6GAP1 A0A1B6DSF1 A0A1B6GW26 A0A0L7QR89 A0A1B6IKU5 E9IIG1 A0A0P6BS44 A0A2J7RDK7 D3TLX8 A0A0P6ITU3 A0A1A9URP2 A0A1A9WLE4 Q16FA7 A0A023ESJ6 Q16ES7 A0A023EVE1 A0A182H850 A0A1B6EIW9 A0A1B0CIG3 B3NHY8 A0A023ET15 B4HJP3 A0A336LK28 B4PKD5 B4QMU7 B0WX08 A0A232FCC4 A0A2S2Q2M4 T1IET8 A0A1Q3FIT4 A0A2P8YWZ2 B4MMG9 Q16PV9 A0A1S4FTF5 A0A2M4BLH7 A0A1W4UGU4 A0A2M4BMC2 A0A3B0KFI0 A0A0M4EDB8 A0A2M4AIN7 A0A2M4AJ04 A0A2M4AIX1 A0A0M4E967 A0A0M4E9Y7 A0A1B0DD06 B3M4H3 A0A0L0C1T2 B4H209 A0A182FDV7
A0A2J7RDJ9 A0A067R8V8 A0A2P8XRY2 A0A2J7RDL5 A0A2J7RDL3 A0A067R6V0 D6WUK9 A0A2J7RDK4 A0A1L8DZQ9 A0A1L8DZV2 E0VT88 A0A158NM28 U4UGG7 N6UIY3 A0A067RID8 F4WHR4 U5EX79 A0A151XHW5 A0A195B4H3 A0A195CX09 A0A154P0U4 A0A151IXB5 A0A088AJX2 A0A151JXQ6 E2B471 A0A2J7RDK5 A0A1B6MC10 A0A1B6KA08 A0A2J7RDK9 E1ZUZ6 E9G9W2 A0A2J7RDK2 A0A1B6JPG7 A0A1A9XB51 A0A310SIY4 A0A0P4YQH5 A0A1B0B6P5 A0A026WG54 A0A1B6DXK1 A0A1B6GTD8 A0A1B6F1J4 A0A1B6GAP1 A0A1B6DSF1 A0A1B6GW26 A0A0L7QR89 A0A1B6IKU5 E9IIG1 A0A0P6BS44 A0A2J7RDK7 D3TLX8 A0A0P6ITU3 A0A1A9URP2 A0A1A9WLE4 Q16FA7 A0A023ESJ6 Q16ES7 A0A023EVE1 A0A182H850 A0A1B6EIW9 A0A1B0CIG3 B3NHY8 A0A023ET15 B4HJP3 A0A336LK28 B4PKD5 B4QMU7 B0WX08 A0A232FCC4 A0A2S2Q2M4 T1IET8 A0A1Q3FIT4 A0A2P8YWZ2 B4MMG9 Q16PV9 A0A1S4FTF5 A0A2M4BLH7 A0A1W4UGU4 A0A2M4BMC2 A0A3B0KFI0 A0A0M4EDB8 A0A2M4AIN7 A0A2M4AJ04 A0A2M4AIX1 A0A0M4E967 A0A0M4E9Y7 A0A1B0DD06 B3M4H3 A0A0L0C1T2 B4H209 A0A182FDV7
PDB
1QWO
E-value=9.41744e-05,
Score=109
Ontologies
PATHWAY
GO
PANTHER
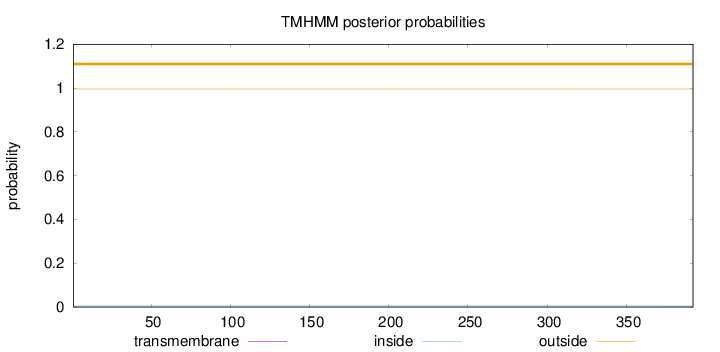
Topology
SignalP
Position: 1 - 16,
Likelihood: 0.998783
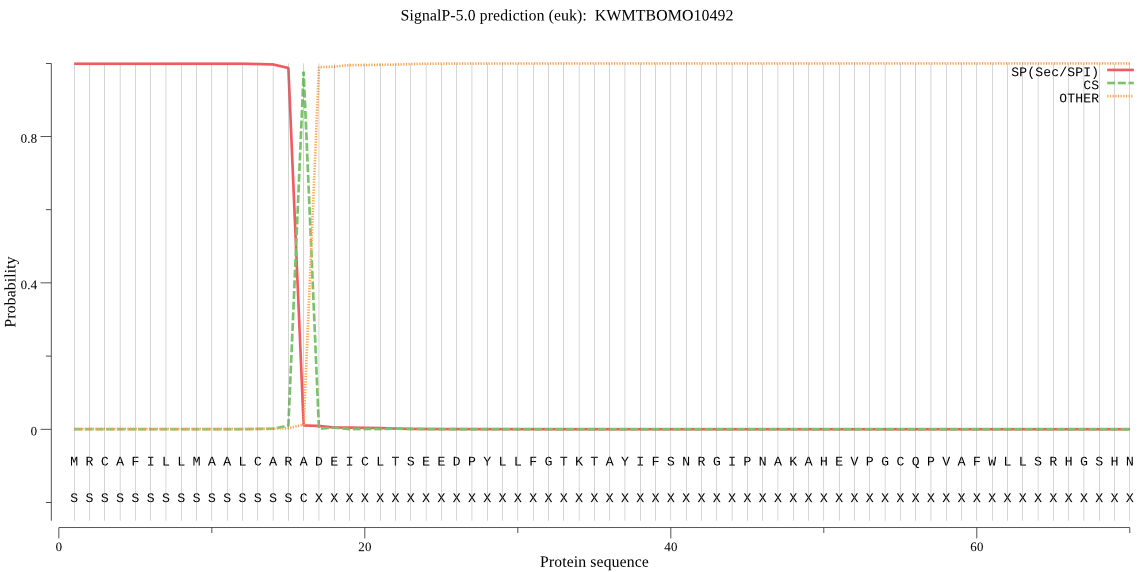
Length:
392
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00544
Exp number, first 60 AAs:
0.00037
Total prob of N-in:
0.00426
outside
1 - 392
Population Genetic Test Statistics
Pi
177.559486
Theta
157.59369
Tajima's D
0.171984
CLR
0.912281
CSRT
0.418479076046198
Interpretation
Uncertain
