Gene
KWMTBOMO10488 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007110
Annotation
small_G_protein_[Spodoptera_exigua]
Full name
Ras-related protein Rac1
Location in the cell
Mitochondrial Reliability : 2.163
Sequence
CDS
ATGGTAGATGGAAAGCCAATCAACTTGGGCCTCTGGGATACAGCCGGACAGGAAGACTACGATCGACTCCGCCCCCTCTCTTACCCACAGACGGACGTCTTCCTTATATGTTTTTCGCTGGTCAACCCAGCCTCATTCGAAAACGTTCGGGCCAAGTGGTATCCAGAAGTTCGGCATCACTGCCCGTCGACGCCCATCATCCTGGTCGGGACCAAGCTCGACTTACGCGAAGACAAGGACACCATTGAAAAGCTCAAGGACAAGAAACTAGCCCCGATCACCTACACGCAGGGTTTGGGCATGTCCAAGGAGATAAACGCTGTCAAGTATTTGGAGTGTTCAGCGCTGACGCAGAAGGGCCTGAAGACCGTGTTCGATGAGGCGATTCGCGCCGTGCTATGCCCGGTGCAGCCCATCAAGGGCCGGCGGCGGCATTACGTCGGAGCCGGTGTGCGCGTCAGAGCTCACACGAACATCCAACAACGCCGCGACACACAGCTAATGCGCCGTCAACCGGACCGCGATACGCACACGCAAACGTTAGCGCGCACCGCCCAACGATACGCGCGACGTAGCGGGGGTCGGTGTGTGTCAAGTCCCTAA
Protein
MVDGKPINLGLWDTAGQEDYDRLRPLSYPQTDVFLICFSLVNPASFENVRAKWYPEVRHHCPSTPIILVGTKLDLREDKDTIEKLKDKKLAPITYTQGLGMSKEINAVKYLECSALTQKGLKTVFDEAIRAVLCPVQPIKGRRRHYVGAGVRVRAHTNIQQRRDTQLMRRQPDRDTHTQTLARTAQRYARRSGGRCVSSP
Summary
Description
Involved in axon outgrowth and myoblast fusion. Plays a role in regulating dorsal closure during embryogenesis. Involved in integrin alpha-PS3/beta-nu-mediated phagocytosis of Gram-positive S.aureus by hemocytes.
Subunit
Interacts with Sra-1. Interacts (via REM 1 repeats) with Pkn (via N-terminus).
Similarity
Belongs to the small GTPase superfamily. Rho family.
Keywords
Cell membrane
Complete proteome
Developmental protein
GTP-binding
Lipoprotein
Membrane
Methylation
Nucleotide-binding
Phagocytosis
Prenylation
Reference proteome
Feature
chain Ras-related protein Rac1
propeptide Removed in mature form
propeptide Removed in mature form
Uniprot
H9JC65
A0A2H1WNP9
J9R033
A0A059VBV6
S4PBQ2
A0A212FC00
+ More
A0A1B6J1H1 A0A1B6GK94 A0A1B6KFE2 A0A1B6DHQ1 A0A1B6JFL9 A0A1B6KIP4 A0A2J7PK11 A0A067RGL0 A0A026WN95 A0A194PPN2 E9JCL1 A0A151J446 F4WFL0 A0A151HYZ1 A0A0J7KSS7 A0A195FT76 E2BW21 A0A158NSH9 E2AAI7 A0A3L8DX32 A0A195CS85 A0A151WI97 V9IKF9 A0A0B4J2M1 A0A194QTP8 A0A069DPS2 A0A224XXI4 A0A0V0GF40 A0A1W4WMW4 A0A0T6B0W7 A0A1Y1N9B9 A0A0P4VSZ1 R4G578 V5GPZ1 A0A2R7WKM6 A0A0A9W7K7 A0A232EZV3 A0A2S2NNV6 A0A1Q3EUE0 A0A1Q3EUF0 D6WGS1 U5EZ98 A0A1J1IBW5 T1DF58 A0A2H8TQX1 A0A0B4J2L1 A0A1L8DKW7 A0A1B0GP55 A0A240SY11 A0A023EIV8 A0A336LVN3 B3M528 E9FRC3 A0A0P5IN29 A0A1W4W6P4 B4LG76 B4L0N4 A0A3B0KG93 Q29EV6 M9PBH7 B4PD66 B4HVR1 B3NB87 B4QLZ5 P40792 V9KJF0 B4N547 B4J2E5 T1H4M0 A0A0P4WCW7 A0A0A7DNX4 Q16V02 A0A3B4DFS4 A0A0P5BPF4 E0VNS7 A0A0M4EC54 A0A1A9UM17 A0A1B0BPU5 A0A1A9ZDN7 A0A1A9WLH6 D3TPD8 A0A3Q2YCX2 A0A165VGD1 A0A0K8TRV5 A0A0L0BT07 A0A3L8SG90 A0A0K8UUK4 A0A034VM04 A0A0A1X180 W8C7L8 A0A0P5DW07 A0A2P4SVU0 A0A0P4WK26
A0A1B6J1H1 A0A1B6GK94 A0A1B6KFE2 A0A1B6DHQ1 A0A1B6JFL9 A0A1B6KIP4 A0A2J7PK11 A0A067RGL0 A0A026WN95 A0A194PPN2 E9JCL1 A0A151J446 F4WFL0 A0A151HYZ1 A0A0J7KSS7 A0A195FT76 E2BW21 A0A158NSH9 E2AAI7 A0A3L8DX32 A0A195CS85 A0A151WI97 V9IKF9 A0A0B4J2M1 A0A194QTP8 A0A069DPS2 A0A224XXI4 A0A0V0GF40 A0A1W4WMW4 A0A0T6B0W7 A0A1Y1N9B9 A0A0P4VSZ1 R4G578 V5GPZ1 A0A2R7WKM6 A0A0A9W7K7 A0A232EZV3 A0A2S2NNV6 A0A1Q3EUE0 A0A1Q3EUF0 D6WGS1 U5EZ98 A0A1J1IBW5 T1DF58 A0A2H8TQX1 A0A0B4J2L1 A0A1L8DKW7 A0A1B0GP55 A0A240SY11 A0A023EIV8 A0A336LVN3 B3M528 E9FRC3 A0A0P5IN29 A0A1W4W6P4 B4LG76 B4L0N4 A0A3B0KG93 Q29EV6 M9PBH7 B4PD66 B4HVR1 B3NB87 B4QLZ5 P40792 V9KJF0 B4N547 B4J2E5 T1H4M0 A0A0P4WCW7 A0A0A7DNX4 Q16V02 A0A3B4DFS4 A0A0P5BPF4 E0VNS7 A0A0M4EC54 A0A1A9UM17 A0A1B0BPU5 A0A1A9ZDN7 A0A1A9WLH6 D3TPD8 A0A3Q2YCX2 A0A165VGD1 A0A0K8TRV5 A0A0L0BT07 A0A3L8SG90 A0A0K8UUK4 A0A034VM04 A0A0A1X180 W8C7L8 A0A0P5DW07 A0A2P4SVU0 A0A0P4WK26
Pubmed
19121390
23660478
24866850
23622113
22118469
24845553
+ More
24508170 26354079 21282665 21719571 20798317 21347285 30249741 26334808 28004739 27129103 25401762 26823975 28648823 18362917 19820115 24330624 24945155 17994087 21292972 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 22936249 7958857 7835340 7720592 12537569 10323867 12818175 17507675 22547074 24402279 26091106 17510324 20566863 20353571 26369729 26108605 30282656 25348373 25830018 24495485
24508170 26354079 21282665 21719571 20798317 21347285 30249741 26334808 28004739 27129103 25401762 26823975 28648823 18362917 19820115 24330624 24945155 17994087 21292972 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 22936249 7958857 7835340 7720592 12537569 10323867 12818175 17507675 22547074 24402279 26091106 17510324 20566863 20353571 26369729 26108605 30282656 25348373 25830018 24495485
EMBL
BABH01014970
BABH01014971
ODYU01009919
SOQ54658.1
JX392403
AFR31806.1
+ More
KJ547699 AHZ89303.1 GAIX01004501 JAA88059.1 AGBW02009256 OWR51248.1 GECU01014694 JAS93012.1 GECZ01025398 GECZ01019608 GECZ01009546 GECZ01006895 JAS44371.1 JAS50161.1 JAS60223.1 JAS62874.1 GEBQ01030073 JAT09904.1 GEDC01012092 JAS25206.1 GECU01009739 JAS97967.1 GEBQ01028664 JAT11313.1 NEVH01024942 PNF16674.1 KK852652 KDR19393.1 KK107161 EZA56574.1 KQ459603 KPI93090.1 GL771848 EFZ09442.1 KQ980211 KYN17208.1 GL888120 EGI66987.1 KQ976719 KYM76748.1 LBMM01003504 KMQ93452.1 KQ981276 KYN43507.1 GL451091 EFN80053.1 ADTU01024932 GL438128 EFN69534.1 QOIP01000003 RLU24793.1 KQ977329 KYN03556.1 KQ983089 KYQ47572.1 JR048909 AEY60799.1 KQ461137 KPJ08842.1 GBGD01003074 JAC85815.1 GFTR01003196 JAW13230.1 GECL01000226 JAP05898.1 LJIG01016368 KRT80811.1 GEZM01012962 JAV92846.1 GDKW01000980 JAI55615.1 ACPB03024203 ACPB03024204 GAHY01000745 JAA76765.1 GALX01004794 JAB63672.1 KK854893 PTY19550.1 GBHO01041136 GBRD01007833 GDHC01017022 GDHC01005035 JAG02468.1 JAG57988.1 JAQ01607.1 JAQ13594.1 NNAY01001454 OXU23922.1 GGMR01006198 MBY18817.1 GFDL01016122 JAV18923.1 GFDL01016123 JAV18922.1 KQ971327 EFA01250.1 GANO01001179 JAB58692.1 CVRI01000047 CRK97775.1 GALA01000772 JAA94080.1 GFXV01004789 MBW16594.1 ABLF02036916 GFDF01007067 JAV07017.1 AJVK01014983 AJWK01000190 AJWK01000191 GAPW01004699 GAPW01004698 JAC08900.1 UFQT01000140 SSX20759.1 CH902618 EDV39507.1 GL732523 EFX90140.1 GDIQ01210423 GDIP01100516 LRGB01002066 JAK41302.1 JAM03199.1 KZS09450.1 CH940647 EDW69384.1 CH933809 EDW18111.1 OUUW01000009 SPP84756.1 CH379070 EAL29953.1 AE014296 AGB93942.1 CM000159 EDW92814.1 CH480817 EDW50026.1 CH954178 EDV50140.1 CM000363 CM002912 EDX08781.1 KMY96784.1 U11823 L38309 Z35642 AY060408 JW865573 AFO98090.1 CH964101 EDW79486.1 CH916366 EDV96004.1 CAQQ02374471 GDRN01049507 JAI66549.1 KF907314 AIU99814.1 CH477607 EAT38364.1 GDIP01185655 JAJ37747.1 DS235346 EEB15033.1 CP012525 ALC42946.1 JXJN01018235 CCAG010005268 EZ423290 ADD19566.1 KT984816 AMZ00368.1 GDAI01000712 JAI16891.1 JRES01001407 KNC23156.1 QUSF01000022 RLW01441.1 GDHF01021972 JAI30342.1 GAKP01014601 JAC44351.1 GBXI01009641 JAD04651.1 GAMC01003544 GAMC01003543 JAC03012.1 GDIP01150411 JAJ72991.1 PPHD01020390 POI28203.1 GDRN01049506 JAI66550.1
KJ547699 AHZ89303.1 GAIX01004501 JAA88059.1 AGBW02009256 OWR51248.1 GECU01014694 JAS93012.1 GECZ01025398 GECZ01019608 GECZ01009546 GECZ01006895 JAS44371.1 JAS50161.1 JAS60223.1 JAS62874.1 GEBQ01030073 JAT09904.1 GEDC01012092 JAS25206.1 GECU01009739 JAS97967.1 GEBQ01028664 JAT11313.1 NEVH01024942 PNF16674.1 KK852652 KDR19393.1 KK107161 EZA56574.1 KQ459603 KPI93090.1 GL771848 EFZ09442.1 KQ980211 KYN17208.1 GL888120 EGI66987.1 KQ976719 KYM76748.1 LBMM01003504 KMQ93452.1 KQ981276 KYN43507.1 GL451091 EFN80053.1 ADTU01024932 GL438128 EFN69534.1 QOIP01000003 RLU24793.1 KQ977329 KYN03556.1 KQ983089 KYQ47572.1 JR048909 AEY60799.1 KQ461137 KPJ08842.1 GBGD01003074 JAC85815.1 GFTR01003196 JAW13230.1 GECL01000226 JAP05898.1 LJIG01016368 KRT80811.1 GEZM01012962 JAV92846.1 GDKW01000980 JAI55615.1 ACPB03024203 ACPB03024204 GAHY01000745 JAA76765.1 GALX01004794 JAB63672.1 KK854893 PTY19550.1 GBHO01041136 GBRD01007833 GDHC01017022 GDHC01005035 JAG02468.1 JAG57988.1 JAQ01607.1 JAQ13594.1 NNAY01001454 OXU23922.1 GGMR01006198 MBY18817.1 GFDL01016122 JAV18923.1 GFDL01016123 JAV18922.1 KQ971327 EFA01250.1 GANO01001179 JAB58692.1 CVRI01000047 CRK97775.1 GALA01000772 JAA94080.1 GFXV01004789 MBW16594.1 ABLF02036916 GFDF01007067 JAV07017.1 AJVK01014983 AJWK01000190 AJWK01000191 GAPW01004699 GAPW01004698 JAC08900.1 UFQT01000140 SSX20759.1 CH902618 EDV39507.1 GL732523 EFX90140.1 GDIQ01210423 GDIP01100516 LRGB01002066 JAK41302.1 JAM03199.1 KZS09450.1 CH940647 EDW69384.1 CH933809 EDW18111.1 OUUW01000009 SPP84756.1 CH379070 EAL29953.1 AE014296 AGB93942.1 CM000159 EDW92814.1 CH480817 EDW50026.1 CH954178 EDV50140.1 CM000363 CM002912 EDX08781.1 KMY96784.1 U11823 L38309 Z35642 AY060408 JW865573 AFO98090.1 CH964101 EDW79486.1 CH916366 EDV96004.1 CAQQ02374471 GDRN01049507 JAI66549.1 KF907314 AIU99814.1 CH477607 EAT38364.1 GDIP01185655 JAJ37747.1 DS235346 EEB15033.1 CP012525 ALC42946.1 JXJN01018235 CCAG010005268 EZ423290 ADD19566.1 KT984816 AMZ00368.1 GDAI01000712 JAI16891.1 JRES01001407 KNC23156.1 QUSF01000022 RLW01441.1 GDHF01021972 JAI30342.1 GAKP01014601 JAC44351.1 GBXI01009641 JAD04651.1 GAMC01003544 GAMC01003543 JAC03012.1 GDIP01150411 JAJ72991.1 PPHD01020390 POI28203.1 GDRN01049506 JAI66550.1
Proteomes
UP000005204
UP000007151
UP000235965
UP000027135
UP000053097
UP000053268
+ More
UP000078492 UP000007755 UP000078540 UP000036403 UP000078541 UP000008237 UP000005205 UP000000311 UP000279307 UP000078542 UP000075809 UP000005203 UP000053240 UP000192223 UP000015103 UP000215335 UP000007266 UP000183832 UP000007819 UP000092462 UP000092461 UP000007801 UP000000305 UP000076858 UP000192221 UP000008792 UP000009192 UP000268350 UP000001819 UP000000803 UP000002282 UP000001292 UP000008711 UP000000304 UP000007798 UP000001070 UP000015102 UP000079169 UP000008820 UP000261440 UP000009046 UP000092553 UP000078200 UP000092460 UP000092445 UP000091820 UP000092444 UP000264820 UP000037069 UP000276834
UP000078492 UP000007755 UP000078540 UP000036403 UP000078541 UP000008237 UP000005205 UP000000311 UP000279307 UP000078542 UP000075809 UP000005203 UP000053240 UP000192223 UP000015103 UP000215335 UP000007266 UP000183832 UP000007819 UP000092462 UP000092461 UP000007801 UP000000305 UP000076858 UP000192221 UP000008792 UP000009192 UP000268350 UP000001819 UP000000803 UP000002282 UP000001292 UP000008711 UP000000304 UP000007798 UP000001070 UP000015102 UP000079169 UP000008820 UP000261440 UP000009046 UP000092553 UP000078200 UP000092460 UP000092445 UP000091820 UP000092444 UP000264820 UP000037069 UP000276834
Pfam
PF00071 Ras
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
H9JC65
A0A2H1WNP9
J9R033
A0A059VBV6
S4PBQ2
A0A212FC00
+ More
A0A1B6J1H1 A0A1B6GK94 A0A1B6KFE2 A0A1B6DHQ1 A0A1B6JFL9 A0A1B6KIP4 A0A2J7PK11 A0A067RGL0 A0A026WN95 A0A194PPN2 E9JCL1 A0A151J446 F4WFL0 A0A151HYZ1 A0A0J7KSS7 A0A195FT76 E2BW21 A0A158NSH9 E2AAI7 A0A3L8DX32 A0A195CS85 A0A151WI97 V9IKF9 A0A0B4J2M1 A0A194QTP8 A0A069DPS2 A0A224XXI4 A0A0V0GF40 A0A1W4WMW4 A0A0T6B0W7 A0A1Y1N9B9 A0A0P4VSZ1 R4G578 V5GPZ1 A0A2R7WKM6 A0A0A9W7K7 A0A232EZV3 A0A2S2NNV6 A0A1Q3EUE0 A0A1Q3EUF0 D6WGS1 U5EZ98 A0A1J1IBW5 T1DF58 A0A2H8TQX1 A0A0B4J2L1 A0A1L8DKW7 A0A1B0GP55 A0A240SY11 A0A023EIV8 A0A336LVN3 B3M528 E9FRC3 A0A0P5IN29 A0A1W4W6P4 B4LG76 B4L0N4 A0A3B0KG93 Q29EV6 M9PBH7 B4PD66 B4HVR1 B3NB87 B4QLZ5 P40792 V9KJF0 B4N547 B4J2E5 T1H4M0 A0A0P4WCW7 A0A0A7DNX4 Q16V02 A0A3B4DFS4 A0A0P5BPF4 E0VNS7 A0A0M4EC54 A0A1A9UM17 A0A1B0BPU5 A0A1A9ZDN7 A0A1A9WLH6 D3TPD8 A0A3Q2YCX2 A0A165VGD1 A0A0K8TRV5 A0A0L0BT07 A0A3L8SG90 A0A0K8UUK4 A0A034VM04 A0A0A1X180 W8C7L8 A0A0P5DW07 A0A2P4SVU0 A0A0P4WK26
A0A1B6J1H1 A0A1B6GK94 A0A1B6KFE2 A0A1B6DHQ1 A0A1B6JFL9 A0A1B6KIP4 A0A2J7PK11 A0A067RGL0 A0A026WN95 A0A194PPN2 E9JCL1 A0A151J446 F4WFL0 A0A151HYZ1 A0A0J7KSS7 A0A195FT76 E2BW21 A0A158NSH9 E2AAI7 A0A3L8DX32 A0A195CS85 A0A151WI97 V9IKF9 A0A0B4J2M1 A0A194QTP8 A0A069DPS2 A0A224XXI4 A0A0V0GF40 A0A1W4WMW4 A0A0T6B0W7 A0A1Y1N9B9 A0A0P4VSZ1 R4G578 V5GPZ1 A0A2R7WKM6 A0A0A9W7K7 A0A232EZV3 A0A2S2NNV6 A0A1Q3EUE0 A0A1Q3EUF0 D6WGS1 U5EZ98 A0A1J1IBW5 T1DF58 A0A2H8TQX1 A0A0B4J2L1 A0A1L8DKW7 A0A1B0GP55 A0A240SY11 A0A023EIV8 A0A336LVN3 B3M528 E9FRC3 A0A0P5IN29 A0A1W4W6P4 B4LG76 B4L0N4 A0A3B0KG93 Q29EV6 M9PBH7 B4PD66 B4HVR1 B3NB87 B4QLZ5 P40792 V9KJF0 B4N547 B4J2E5 T1H4M0 A0A0P4WCW7 A0A0A7DNX4 Q16V02 A0A3B4DFS4 A0A0P5BPF4 E0VNS7 A0A0M4EC54 A0A1A9UM17 A0A1B0BPU5 A0A1A9ZDN7 A0A1A9WLH6 D3TPD8 A0A3Q2YCX2 A0A165VGD1 A0A0K8TRV5 A0A0L0BT07 A0A3L8SG90 A0A0K8UUK4 A0A034VM04 A0A0A1X180 W8C7L8 A0A0P5DW07 A0A2P4SVU0 A0A0P4WK26
PDB
5FI0
E-value=1.09891e-73,
Score=700
Ontologies
KEGG
PATHWAY
GO
GO:0005525
GO:0003924
GO:0007264
GO:0016021
GO:0016477
GO:0007266
GO:0008045
GO:0030036
GO:0005737
GO:0019901
GO:0031410
GO:0042995
GO:0005886
GO:0046843
GO:0016028
GO:0007394
GO:0019897
GO:0007419
GO:0007413
GO:0032147
GO:0050975
GO:0031532
GO:1902669
GO:0035011
GO:0097206
GO:0032794
GO:0035212
GO:0090303
GO:0007390
GO:0048010
GO:0007395
GO:0048814
GO:0010593
GO:0051491
GO:0007516
GO:0001954
GO:0007520
GO:0071902
GO:0009611
GO:0042052
GO:0045773
GO:0008258
GO:0008078
GO:0048615
GO:0035320
GO:0048675
GO:0051450
GO:0007298
GO:0007422
GO:1904059
GO:0030866
GO:0016199
GO:0030032
GO:0007613
GO:0071907
GO:0048747
GO:1903688
GO:0001726
GO:0007254
GO:0051017
GO:0007435
GO:0051963
GO:0008347
GO:0035099
GO:0016203
GO:0046664
GO:0016358
GO:0046330
GO:0007426
GO:0002433
GO:0034334
GO:0007411
GO:0008283
GO:0007391
GO:0030707
GO:0000902
GO:0007015
GO:0048813
GO:0005829
GO:0050770
GO:0048812
GO:0031175
GO:0050807
GO:0035010
GO:0007424
GO:0008237
GO:0005245
GO:0005891
GO:0004674
GO:0006418
GO:0003723
GO:0046872
GO:0003824
Topology
Subcellular location
Cell membrane
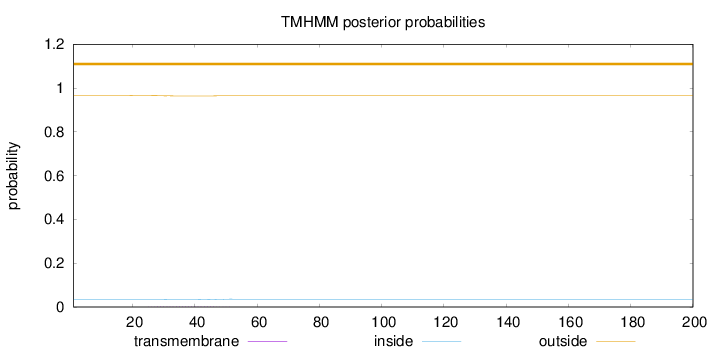
Length:
200
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0422
Exp number, first 60 AAs:
0.0422
Total prob of N-in:
0.03372
outside
1 - 200
Population Genetic Test Statistics
Pi
213.017444
Theta
165.950648
Tajima's D
0.72361
CLR
0.380542
CSRT
0.581720913954302
Interpretation
Uncertain
