Gene
KWMTBOMO10479
Annotation
PREDICTED:_uncharacterized_protein_LOC106133666_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 1.992
Sequence
CDS
ATGTTTTGTGATTTTGGTGGTTCTTCAGACTTCACGCCAATCGCCTTAGATGGTGTATCCCTGCCAGTCTGCTCCGACTTCAAGTACCTCGGCTCTATACTCCAGAACGACGGCAACATAGACCGGGACGTCACAAATAGAATAAACGCTGGTTGGATGAAATGGCGACAGGTCTCTGCAACAGCATGCGATCGTAGAATGCCCCTTCAGCTCAAGGGCAAAATTTACAAAACGATCATTAGACCTGTCGTGCTGTATGGATCTGAGTGTTGGGCGACGAAAGTTAAGCATGAAAGAGGAGTGCATGCGGCAGAGATGCGAATGTTGAGATGGTTGTGTGGAGTAACAAGAATGGATAAGATAAAGAATAAGTATATCAGAGGAAGTGTGAAAGTGGCCCCGGTAAACAACAAATTAAGGAGCGGACGGTTAGCGTGGTATGGACATGTGATGCGTAGAGAGAAGATGCATGTGACTAGGAGATGTATGGAAATGGTAGTGCAAGGTAGAGGGGGAAGAGGTCGACCGAAGAAGACATGGATGGAGTGTGTGAATAACGATATGAGAGAGAGAGGAGTGAGTGTTGAGATGACGGCTGATAGAAGAGAATGGAAGAGAAAAATTAGTTGTGCCGACCCCACATAG
Protein
MFCDFGGSSDFTPIALDGVSLPVCSDFKYLGSILQNDGNIDRDVTNRINAGWMKWRQVSATACDRRMPLQLKGKIYKTIIRPVVLYGSECWATKVKHERGVHAAEMRMLRWLCGVTRMDKIKNKYIRGSVKVAPVNNKLRSGRLAWYGHVMRREKMHVTRRCMEMVVQGRGGRGRPKKTWMECVNNDMRERGVSVEMTADRREWKRKISCADPT
Summary
Uniprot
A0A2A4IZ11
A0A2A4JTH5
A0A016SG79
A0A016SHD2
A0A2I0A7B2
A0A2I0BDY2
+ More
A0A016SBS5 A0A016TH37 A0A016SDV3 A0A2I0BFB2 A0A2I0A8G9 A0A016T4Z4 A0A016VL18 A0A2I0A8E3 A0A016TU56 A0A016SKJ5 A0A016TV72 A0A3S4QVV5 A0A2I0A4R0 A0A016S415 A0A016WRX0 A0A016UZ62 A0A151UDZ2 A0A016SKB8 A0A2I0X0R9 A0A2I0XCV6 A0A016T1D4 A0A016SQ37 A0A016T3U9 A0A016VC06 A0A2I0WD15 A0A016UAZ5 Q1AKH8 A0A016S5A7 A0A016X4X8 A0A2I0VW23 A0A016WMG6 A0A016TB11 A0A016UUZ0 A0A2I0WE43 A0A2H9ZWL9 A0A1D6N4Q8 A0A1D6GX82 A0A016VGS9 A0A016X4U3 A0A1D6Q287 A0A1D6EFS5 A0A2I0VKF6 A0A016V2A1 A0A2S3HU98 A0A016X2L9 A0A016TBY7 A0A016U5E2 A0A2I0W1V0 A0A016WTX6 A0A2I0XFV9 A0A0V0IX54 A0A2I0VI73 A0A2I0X0N4 A0A2I0X2W5 A0A016WBG8 A0A2T8I3I2 A0A016VG09 A0A016WAY5 A0A1D6GN15 A0A2I0X0J0 A0A2S3HUX6 A0A016X2P5 D7F169 C4JB60 A0A2I0V6Y4 C6Y4D5 A0A3L6FA92 A0A1D6DWM6 A0A3L6FFF5 A0A016SQS9 A0A2T8IB31 A0A016TVA5 A0A2T7F8M8 A0A1D6HQE7 A0A016S355 A0A1U7X9E6 A0A2I0WGV1 A0A3L6EUT5 A0A3L6DYM7 A0A1D6GGM1 A0A016VX07 A0A2T7F873 A0A2S3IIQ5 A0A016S7U7 A0A016TJ58 A0A016W9G0 A0A3L6FU31 A0A016X2X2 A0A1D6F347 A0A016RXP6 A0A016UIA0 A0A2T8KNP5 M1BPI1 A0A016WIE8
A0A016SBS5 A0A016TH37 A0A016SDV3 A0A2I0BFB2 A0A2I0A8G9 A0A016T4Z4 A0A016VL18 A0A2I0A8E3 A0A016TU56 A0A016SKJ5 A0A016TV72 A0A3S4QVV5 A0A2I0A4R0 A0A016S415 A0A016WRX0 A0A016UZ62 A0A151UDZ2 A0A016SKB8 A0A2I0X0R9 A0A2I0XCV6 A0A016T1D4 A0A016SQ37 A0A016T3U9 A0A016VC06 A0A2I0WD15 A0A016UAZ5 Q1AKH8 A0A016S5A7 A0A016X4X8 A0A2I0VW23 A0A016WMG6 A0A016TB11 A0A016UUZ0 A0A2I0WE43 A0A2H9ZWL9 A0A1D6N4Q8 A0A1D6GX82 A0A016VGS9 A0A016X4U3 A0A1D6Q287 A0A1D6EFS5 A0A2I0VKF6 A0A016V2A1 A0A2S3HU98 A0A016X2L9 A0A016TBY7 A0A016U5E2 A0A2I0W1V0 A0A016WTX6 A0A2I0XFV9 A0A0V0IX54 A0A2I0VI73 A0A2I0X0N4 A0A2I0X2W5 A0A016WBG8 A0A2T8I3I2 A0A016VG09 A0A016WAY5 A0A1D6GN15 A0A2I0X0J0 A0A2S3HUX6 A0A016X2P5 D7F169 C4JB60 A0A2I0V6Y4 C6Y4D5 A0A3L6FA92 A0A1D6DWM6 A0A3L6FFF5 A0A016SQS9 A0A2T8IB31 A0A016TVA5 A0A2T7F8M8 A0A1D6HQE7 A0A016S355 A0A1U7X9E6 A0A2I0WGV1 A0A3L6EUT5 A0A3L6DYM7 A0A1D6GGM1 A0A016VX07 A0A2T7F873 A0A2S3IIQ5 A0A016S7U7 A0A016TJ58 A0A016W9G0 A0A3L6FU31 A0A016X2X2 A0A1D6F347 A0A016RXP6 A0A016UIA0 A0A2T8KNP5 M1BPI1 A0A016WIE8
Pubmed
EMBL
NWSH01004369
PCG65177.1
NWSH01000612
PCG75311.1
JARK01001564
EYB89718.1
+ More
EYB89719.1 KZ452013 PKA51430.1 KZ451888 PKA66013.1 JARK01001594 EYB87754.1 JARK01001437 EYC02289.1 JARK01001582 EYB88469.1 KZ451885 PKA66495.1 KZ452012 PKA51832.1 JARK01001473 EYB97757.1 JARK01001345 EYC27463.1 PKA51807.1 JARK01001413 EYC06291.1 JARK01001546 EYB91135.1 EYC06288.1 NCKU01002988 RWS08398.1 KZ452023 PKA50536.1 JARK01001642 EYB85037.1 JARK01000157 EYC41778.1 JARK01001358 EYC20465.1 AGCT01024946 KYP77525.1 EYB91138.1 KZ502235 PKU81499.1 KZ501974 PKU85733.1 JARK01001486 EYB96487.1 JARK01001529 EYB92481.1 JARK01001478 EYB97301.1 JARK01001348 EYC24955.1 KZ502741 PKU73550.1 JARK01001383 EYC12479.1 DQ070849 ABB00038.1 JARK01001626 EYB85828.1 JARK01000002 EYC46303.1 KZ503176 PKU67611.1 JARK01000211 EYC40457.1 JARK01001455 EYB99857.1 JARK01001362 EYC18796.1 KZ502716 PKU73925.1 KZ453102 PKA47709.1 CM007649 ONM35621.1 CM000781 AQK66095.1 AQK67441.1 JARK01001346 EYC26232.1 EYC46273.1 CM000780 CM007650 AQK52712.1 ONM54013.1 CM007648 ONM19028.1 KZ503459 PKU63885.1 JARK01001356 EYC20868.1 CM008050 PAN29912.1 JARK01000001 EYC46334.1 JARK01001453 EYC00158.1 JARK01001391 EYC10519.1 KZ503020 PKU69639.1 JARK01000103 EYC43100.1 KZ502732 KZ501922 KZ501830 PKU73666.1 PKU86798.1 PKU87987.1 GEDG01001285 JAP37101.1 KZ503527 PKU63110.1 PKU81488.1 KZ502564 KZ502519 KZ502203 PKU76010.1 PKU77283.1 PKU82246.1 JARK01000474 EYC36617.1 CM008054 PVH32202.1 EYC26231.1 JARK01000458 EYC36771.1 AQK64650.1 KZ502242 PKU81438.1 PAN30387.1 JARK01000005 EYC46180.1 FJ265554 ADI61822.1 BT088057 ACR38410.1 KZ504148 PKU59166.1 CU462842 CBA11992.1 NCVQ01000005 PWZ29813.1 ONM13079.1 NCVQ01000004 PWZ31071.1 JARK01001524 EYB92930.1 CM008052 PVH34896.1 JARK01001410 EYC06740.1 CM009749 PUZ76420.1 AQK76494.1 JARK01000189 EYB85038.1 EYC40926.1 KZ502668 PKU74878.1 NCVQ01000006 PWZ23087.1 NCVQ01000008 PWZ13007.1 AQK62700.1 JARK01001340 EYC31308.1 PUZ76291.1 PAN45362.2 JARK01001617 EYB86334.1 JARK01001432 EYC02999.1 JARK01000517 EYC36246.1 NCVQ01000003 PWZ38398.1 EYC46181.1 ONM25835.1 JARK01001678 EYB83113.1 JARK01001374 EYC15074.1 CM008047 PVH63795.1 JARK01000257 EYC39401.1
EYB89719.1 KZ452013 PKA51430.1 KZ451888 PKA66013.1 JARK01001594 EYB87754.1 JARK01001437 EYC02289.1 JARK01001582 EYB88469.1 KZ451885 PKA66495.1 KZ452012 PKA51832.1 JARK01001473 EYB97757.1 JARK01001345 EYC27463.1 PKA51807.1 JARK01001413 EYC06291.1 JARK01001546 EYB91135.1 EYC06288.1 NCKU01002988 RWS08398.1 KZ452023 PKA50536.1 JARK01001642 EYB85037.1 JARK01000157 EYC41778.1 JARK01001358 EYC20465.1 AGCT01024946 KYP77525.1 EYB91138.1 KZ502235 PKU81499.1 KZ501974 PKU85733.1 JARK01001486 EYB96487.1 JARK01001529 EYB92481.1 JARK01001478 EYB97301.1 JARK01001348 EYC24955.1 KZ502741 PKU73550.1 JARK01001383 EYC12479.1 DQ070849 ABB00038.1 JARK01001626 EYB85828.1 JARK01000002 EYC46303.1 KZ503176 PKU67611.1 JARK01000211 EYC40457.1 JARK01001455 EYB99857.1 JARK01001362 EYC18796.1 KZ502716 PKU73925.1 KZ453102 PKA47709.1 CM007649 ONM35621.1 CM000781 AQK66095.1 AQK67441.1 JARK01001346 EYC26232.1 EYC46273.1 CM000780 CM007650 AQK52712.1 ONM54013.1 CM007648 ONM19028.1 KZ503459 PKU63885.1 JARK01001356 EYC20868.1 CM008050 PAN29912.1 JARK01000001 EYC46334.1 JARK01001453 EYC00158.1 JARK01001391 EYC10519.1 KZ503020 PKU69639.1 JARK01000103 EYC43100.1 KZ502732 KZ501922 KZ501830 PKU73666.1 PKU86798.1 PKU87987.1 GEDG01001285 JAP37101.1 KZ503527 PKU63110.1 PKU81488.1 KZ502564 KZ502519 KZ502203 PKU76010.1 PKU77283.1 PKU82246.1 JARK01000474 EYC36617.1 CM008054 PVH32202.1 EYC26231.1 JARK01000458 EYC36771.1 AQK64650.1 KZ502242 PKU81438.1 PAN30387.1 JARK01000005 EYC46180.1 FJ265554 ADI61822.1 BT088057 ACR38410.1 KZ504148 PKU59166.1 CU462842 CBA11992.1 NCVQ01000005 PWZ29813.1 ONM13079.1 NCVQ01000004 PWZ31071.1 JARK01001524 EYB92930.1 CM008052 PVH34896.1 JARK01001410 EYC06740.1 CM009749 PUZ76420.1 AQK76494.1 JARK01000189 EYB85038.1 EYC40926.1 KZ502668 PKU74878.1 NCVQ01000006 PWZ23087.1 NCVQ01000008 PWZ13007.1 AQK62700.1 JARK01001340 EYC31308.1 PUZ76291.1 PAN45362.2 JARK01001617 EYB86334.1 JARK01001432 EYC02999.1 JARK01000517 EYC36246.1 NCVQ01000003 PWZ38398.1 EYC46181.1 ONM25835.1 JARK01001678 EYB83113.1 JARK01001374 EYC15074.1 CM008047 PVH63795.1 JARK01000257 EYC39401.1
Proteomes
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A2A4IZ11
A0A2A4JTH5
A0A016SG79
A0A016SHD2
A0A2I0A7B2
A0A2I0BDY2
+ More
A0A016SBS5 A0A016TH37 A0A016SDV3 A0A2I0BFB2 A0A2I0A8G9 A0A016T4Z4 A0A016VL18 A0A2I0A8E3 A0A016TU56 A0A016SKJ5 A0A016TV72 A0A3S4QVV5 A0A2I0A4R0 A0A016S415 A0A016WRX0 A0A016UZ62 A0A151UDZ2 A0A016SKB8 A0A2I0X0R9 A0A2I0XCV6 A0A016T1D4 A0A016SQ37 A0A016T3U9 A0A016VC06 A0A2I0WD15 A0A016UAZ5 Q1AKH8 A0A016S5A7 A0A016X4X8 A0A2I0VW23 A0A016WMG6 A0A016TB11 A0A016UUZ0 A0A2I0WE43 A0A2H9ZWL9 A0A1D6N4Q8 A0A1D6GX82 A0A016VGS9 A0A016X4U3 A0A1D6Q287 A0A1D6EFS5 A0A2I0VKF6 A0A016V2A1 A0A2S3HU98 A0A016X2L9 A0A016TBY7 A0A016U5E2 A0A2I0W1V0 A0A016WTX6 A0A2I0XFV9 A0A0V0IX54 A0A2I0VI73 A0A2I0X0N4 A0A2I0X2W5 A0A016WBG8 A0A2T8I3I2 A0A016VG09 A0A016WAY5 A0A1D6GN15 A0A2I0X0J0 A0A2S3HUX6 A0A016X2P5 D7F169 C4JB60 A0A2I0V6Y4 C6Y4D5 A0A3L6FA92 A0A1D6DWM6 A0A3L6FFF5 A0A016SQS9 A0A2T8IB31 A0A016TVA5 A0A2T7F8M8 A0A1D6HQE7 A0A016S355 A0A1U7X9E6 A0A2I0WGV1 A0A3L6EUT5 A0A3L6DYM7 A0A1D6GGM1 A0A016VX07 A0A2T7F873 A0A2S3IIQ5 A0A016S7U7 A0A016TJ58 A0A016W9G0 A0A3L6FU31 A0A016X2X2 A0A1D6F347 A0A016RXP6 A0A016UIA0 A0A2T8KNP5 M1BPI1 A0A016WIE8
A0A016SBS5 A0A016TH37 A0A016SDV3 A0A2I0BFB2 A0A2I0A8G9 A0A016T4Z4 A0A016VL18 A0A2I0A8E3 A0A016TU56 A0A016SKJ5 A0A016TV72 A0A3S4QVV5 A0A2I0A4R0 A0A016S415 A0A016WRX0 A0A016UZ62 A0A151UDZ2 A0A016SKB8 A0A2I0X0R9 A0A2I0XCV6 A0A016T1D4 A0A016SQ37 A0A016T3U9 A0A016VC06 A0A2I0WD15 A0A016UAZ5 Q1AKH8 A0A016S5A7 A0A016X4X8 A0A2I0VW23 A0A016WMG6 A0A016TB11 A0A016UUZ0 A0A2I0WE43 A0A2H9ZWL9 A0A1D6N4Q8 A0A1D6GX82 A0A016VGS9 A0A016X4U3 A0A1D6Q287 A0A1D6EFS5 A0A2I0VKF6 A0A016V2A1 A0A2S3HU98 A0A016X2L9 A0A016TBY7 A0A016U5E2 A0A2I0W1V0 A0A016WTX6 A0A2I0XFV9 A0A0V0IX54 A0A2I0VI73 A0A2I0X0N4 A0A2I0X2W5 A0A016WBG8 A0A2T8I3I2 A0A016VG09 A0A016WAY5 A0A1D6GN15 A0A2I0X0J0 A0A2S3HUX6 A0A016X2P5 D7F169 C4JB60 A0A2I0V6Y4 C6Y4D5 A0A3L6FA92 A0A1D6DWM6 A0A3L6FFF5 A0A016SQS9 A0A2T8IB31 A0A016TVA5 A0A2T7F8M8 A0A1D6HQE7 A0A016S355 A0A1U7X9E6 A0A2I0WGV1 A0A3L6EUT5 A0A3L6DYM7 A0A1D6GGM1 A0A016VX07 A0A2T7F873 A0A2S3IIQ5 A0A016S7U7 A0A016TJ58 A0A016W9G0 A0A3L6FU31 A0A016X2X2 A0A1D6F347 A0A016RXP6 A0A016UIA0 A0A2T8KNP5 M1BPI1 A0A016WIE8
Ontologies
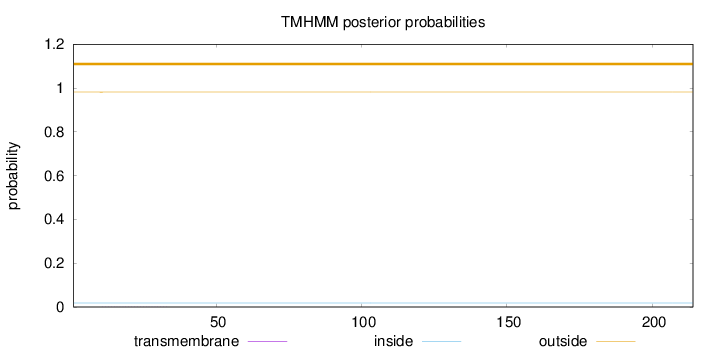
Topology
Length:
214
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00582
Exp number, first 60 AAs:
0.00546
Total prob of N-in:
0.01836
outside
1 - 214
Population Genetic Test Statistics
Pi
233.461053
Theta
162.443076
Tajima's D
1.403685
CLR
0.337228
CSRT
0.763911804409779
Interpretation
Uncertain
