Gene
KWMTBOMO10468
Pre Gene Modal
BGIBMGA007102
Annotation
PREDICTED:_uncharacterized_protein_LOC106118897_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 4.761
Sequence
CDS
ATGTTATGCGCTGCCATCATGACAGACCACTGGGAGCACGTGGCGTGGGACCGAGCCGCGCTGACCGCGCTAGCTAACACGTCGCACATACCGCTGCAGTGGCTGTTAGATGAAAAAGTTGTCAAAGTCGACGACCCGAATAACAGAAAAGGAATCGGGACCTTTTTGGTCCCTATGAACGGAGGTATCTGGACGATGTGCGTCTCTTTAGAAGAGGAGGAAGCCACGATTTTGTCAAGAGTTGGCTTCCCAAAGGAACCGGTCTGCACGAACTATTTGGCCGATAGCGACTACGAAGAGCCCCGGGCGGATTGGCAGCACCGTATGCAAAATCTATCGATATCTTGTGCGTTGGTCTGCCTGATAGTGCTGGGAAGCGCGGCGCTGGTGGGCGCTTTCGGAGTCTGCAAGCATCAGATCAGCGCCGTTCTTATAACCGGAGTTATGTATTTATTGGCCGGTCTCTTTGCAATGTTTACGTTAATGATAATCCACTTCAAACGCGTCCAGCGGGTCTCGTCCCGGGGTAGTTCTCATGGAGACGACACGGGAGATGGAGTAGTCGGTCCCAAGACTACAGTCATTCCTCTCCTTTCCGCAAGAGAATTCTCCACGTCGTGGTCCCTCGAACTCGGCTGGGGGGGAGTTGTACTAGCGACTACAACTTCACTACTGTGGATATTACTATCGAAAATAATGAGATACAATCCCATATCGACTATGCTGTTGTAA
Protein
MLCAAIMTDHWEHVAWDRAALTALANTSHIPLQWLLDEKVVKVDDPNNRKGIGTFLVPMNGGIWTMCVSLEEEEATILSRVGFPKEPVCTNYLADSDYEEPRADWQHRMQNLSISCALVCLIVLGSAALVGAFGVCKHQISAVLITGVMYLLAGLFAMFTLMIIHFKRVQRVSSRGSSHGDDTGDGVVGPKTTVIPLLSAREFSTSWSLELGWGGVVLATTTSLLWILLSKIMRYNPISTMLL
Summary
Uniprot
A0A194PPM3
A0A3S2LQA4
A0A212ETY3
A0A2H1VIW5
A0A194QTN8
D2A551
+ More
A0A1Y1JUK8 U4U5J5 N6SU50 A0A1W4XLR7 A0A182IJG2 A0A084W354 A0A182HER6 W8B6V2 A0A182QH12 A0A182RPE8 A0A1I8PXM2 A0A1W4XKW3 A0A182PDZ7 A0A182M2F8 A0A182HTJ5 A0A182XIS1 A0A1S4FC82 A0A1J1IV45 A0A0M8ZZP9 A0A1W4W382 W8BLV4 A0A1I8N3L3 W8AZV0 A0A182VRU9 A0A310S6N0 A0A1I8PXK3 B4PTV5 A0A2A3EAZ1 A0A088AR78 B4KDB5 A0A1B6DGP9 A0A3B0JSP8 A0A0C9QMI6 A0A182XUX9 Q294J7 A0A2J7QGI5 A0A154P3A5 A0A3L8DIB7 A0A026WTL8 A0A1B0EU37 A0A151WRR7 A0A158NZF9 A0A182F724 A0A182N7B1 A0A0L7REW1 A0A195CSB8 A0A034WUH6 A0A151ISY0 E2AB20 A0A195FY61 E9J723 E2C4L9 A0A067QVU4 A0A182LBW6 A0A182UJB0 A0A0A9WR29 A0A0K8T9V2 A0A224XXA4 A0A0A9WFR9 A0A0K8T9W4 A0A336M2S7 A0A2S2QL02 J9LG32
A0A1Y1JUK8 U4U5J5 N6SU50 A0A1W4XLR7 A0A182IJG2 A0A084W354 A0A182HER6 W8B6V2 A0A182QH12 A0A182RPE8 A0A1I8PXM2 A0A1W4XKW3 A0A182PDZ7 A0A182M2F8 A0A182HTJ5 A0A182XIS1 A0A1S4FC82 A0A1J1IV45 A0A0M8ZZP9 A0A1W4W382 W8BLV4 A0A1I8N3L3 W8AZV0 A0A182VRU9 A0A310S6N0 A0A1I8PXK3 B4PTV5 A0A2A3EAZ1 A0A088AR78 B4KDB5 A0A1B6DGP9 A0A3B0JSP8 A0A0C9QMI6 A0A182XUX9 Q294J7 A0A2J7QGI5 A0A154P3A5 A0A3L8DIB7 A0A026WTL8 A0A1B0EU37 A0A151WRR7 A0A158NZF9 A0A182F724 A0A182N7B1 A0A0L7REW1 A0A195CSB8 A0A034WUH6 A0A151ISY0 E2AB20 A0A195FY61 E9J723 E2C4L9 A0A067QVU4 A0A182LBW6 A0A182UJB0 A0A0A9WR29 A0A0K8T9V2 A0A224XXA4 A0A0A9WFR9 A0A0K8T9W4 A0A336M2S7 A0A2S2QL02 J9LG32
Pubmed
EMBL
KQ459603
KPI93080.1
RSAL01000031
RVE51621.1
AGBW02012517
OWR44953.1
+ More
ODYU01002794 SOQ40736.1 KQ461137 KPJ08832.1 KQ971361 EFA05133.2 GEZM01100786 JAV52793.1 KB632155 ERL89174.1 APGK01056937 KB741277 ENN71219.1 ATLV01019817 KE525279 KFB44648.1 JXUM01005810 JXUM01005811 JXUM01005812 JXUM01005813 JXUM01005814 KQ560202 KXJ83869.1 GAMC01012198 JAB94357.1 AXCN02000146 AXCM01004826 CVRI01000057 CRL02409.1 KQ435791 KOX74400.1 GAMC01012199 JAB94356.1 GAMC01012200 JAB94355.1 KQ767621 OAD53384.1 CM000160 EDW96566.2 KZ288313 PBC28356.1 CH933806 EDW14897.2 GEDC01012435 JAS24863.1 OUUW01000008 SPP84033.1 GBYB01001837 GBYB01001838 JAG71604.1 JAG71605.1 CM000070 EAL28967.4 NEVH01014362 PNF27698.1 KQ434796 KZC05718.1 QOIP01000008 RLU19578.1 KK107109 EZA59357.1 CCAG010000815 KQ982807 KYQ50365.1 ADTU01004588 KQ414608 KOC69368.1 KQ977349 KYN03397.1 GAKP01000955 JAC57997.1 KQ981047 KYN10036.1 GL438234 EFN69296.1 KQ981169 KYN45378.1 GL768421 EFZ11374.1 GL452536 EFN77099.1 KK853186 KDR10065.1 GBHO01036299 JAG07305.1 GBRD01003493 JAG62328.1 GFTR01003795 JAW12631.1 GBHO01036302 JAG07302.1 GBRD01003492 JAG62329.1 UFQT01000375 SSX23701.1 GGMS01009017 MBY78220.1 ABLF02018459
ODYU01002794 SOQ40736.1 KQ461137 KPJ08832.1 KQ971361 EFA05133.2 GEZM01100786 JAV52793.1 KB632155 ERL89174.1 APGK01056937 KB741277 ENN71219.1 ATLV01019817 KE525279 KFB44648.1 JXUM01005810 JXUM01005811 JXUM01005812 JXUM01005813 JXUM01005814 KQ560202 KXJ83869.1 GAMC01012198 JAB94357.1 AXCN02000146 AXCM01004826 CVRI01000057 CRL02409.1 KQ435791 KOX74400.1 GAMC01012199 JAB94356.1 GAMC01012200 JAB94355.1 KQ767621 OAD53384.1 CM000160 EDW96566.2 KZ288313 PBC28356.1 CH933806 EDW14897.2 GEDC01012435 JAS24863.1 OUUW01000008 SPP84033.1 GBYB01001837 GBYB01001838 JAG71604.1 JAG71605.1 CM000070 EAL28967.4 NEVH01014362 PNF27698.1 KQ434796 KZC05718.1 QOIP01000008 RLU19578.1 KK107109 EZA59357.1 CCAG010000815 KQ982807 KYQ50365.1 ADTU01004588 KQ414608 KOC69368.1 KQ977349 KYN03397.1 GAKP01000955 JAC57997.1 KQ981047 KYN10036.1 GL438234 EFN69296.1 KQ981169 KYN45378.1 GL768421 EFZ11374.1 GL452536 EFN77099.1 KK853186 KDR10065.1 GBHO01036299 JAG07305.1 GBRD01003493 JAG62328.1 GFTR01003795 JAW12631.1 GBHO01036302 JAG07302.1 GBRD01003492 JAG62329.1 UFQT01000375 SSX23701.1 GGMS01009017 MBY78220.1 ABLF02018459
Proteomes
UP000053268
UP000283053
UP000007151
UP000053240
UP000007266
UP000030742
+ More
UP000019118 UP000192223 UP000075880 UP000030765 UP000069940 UP000249989 UP000075886 UP000075900 UP000095300 UP000075885 UP000075883 UP000076407 UP000183832 UP000053105 UP000192221 UP000095301 UP000075920 UP000002282 UP000242457 UP000005203 UP000009192 UP000268350 UP000076408 UP000001819 UP000235965 UP000076502 UP000279307 UP000053097 UP000092444 UP000075809 UP000005205 UP000069272 UP000075884 UP000053825 UP000078542 UP000078492 UP000000311 UP000078541 UP000008237 UP000027135 UP000075882 UP000075902 UP000007819
UP000019118 UP000192223 UP000075880 UP000030765 UP000069940 UP000249989 UP000075886 UP000075900 UP000095300 UP000075885 UP000075883 UP000076407 UP000183832 UP000053105 UP000192221 UP000095301 UP000075920 UP000002282 UP000242457 UP000005203 UP000009192 UP000268350 UP000076408 UP000001819 UP000235965 UP000076502 UP000279307 UP000053097 UP000092444 UP000075809 UP000005205 UP000069272 UP000075884 UP000053825 UP000078542 UP000078492 UP000000311 UP000078541 UP000008237 UP000027135 UP000075882 UP000075902 UP000007819
Pfam
PF13903 Claudin_2
Interpro
IPR004031
PMP22/EMP/MP20/Claudin
ProteinModelPortal
A0A194PPM3
A0A3S2LQA4
A0A212ETY3
A0A2H1VIW5
A0A194QTN8
D2A551
+ More
A0A1Y1JUK8 U4U5J5 N6SU50 A0A1W4XLR7 A0A182IJG2 A0A084W354 A0A182HER6 W8B6V2 A0A182QH12 A0A182RPE8 A0A1I8PXM2 A0A1W4XKW3 A0A182PDZ7 A0A182M2F8 A0A182HTJ5 A0A182XIS1 A0A1S4FC82 A0A1J1IV45 A0A0M8ZZP9 A0A1W4W382 W8BLV4 A0A1I8N3L3 W8AZV0 A0A182VRU9 A0A310S6N0 A0A1I8PXK3 B4PTV5 A0A2A3EAZ1 A0A088AR78 B4KDB5 A0A1B6DGP9 A0A3B0JSP8 A0A0C9QMI6 A0A182XUX9 Q294J7 A0A2J7QGI5 A0A154P3A5 A0A3L8DIB7 A0A026WTL8 A0A1B0EU37 A0A151WRR7 A0A158NZF9 A0A182F724 A0A182N7B1 A0A0L7REW1 A0A195CSB8 A0A034WUH6 A0A151ISY0 E2AB20 A0A195FY61 E9J723 E2C4L9 A0A067QVU4 A0A182LBW6 A0A182UJB0 A0A0A9WR29 A0A0K8T9V2 A0A224XXA4 A0A0A9WFR9 A0A0K8T9W4 A0A336M2S7 A0A2S2QL02 J9LG32
A0A1Y1JUK8 U4U5J5 N6SU50 A0A1W4XLR7 A0A182IJG2 A0A084W354 A0A182HER6 W8B6V2 A0A182QH12 A0A182RPE8 A0A1I8PXM2 A0A1W4XKW3 A0A182PDZ7 A0A182M2F8 A0A182HTJ5 A0A182XIS1 A0A1S4FC82 A0A1J1IV45 A0A0M8ZZP9 A0A1W4W382 W8BLV4 A0A1I8N3L3 W8AZV0 A0A182VRU9 A0A310S6N0 A0A1I8PXK3 B4PTV5 A0A2A3EAZ1 A0A088AR78 B4KDB5 A0A1B6DGP9 A0A3B0JSP8 A0A0C9QMI6 A0A182XUX9 Q294J7 A0A2J7QGI5 A0A154P3A5 A0A3L8DIB7 A0A026WTL8 A0A1B0EU37 A0A151WRR7 A0A158NZF9 A0A182F724 A0A182N7B1 A0A0L7REW1 A0A195CSB8 A0A034WUH6 A0A151ISY0 E2AB20 A0A195FY61 E9J723 E2C4L9 A0A067QVU4 A0A182LBW6 A0A182UJB0 A0A0A9WR29 A0A0K8T9V2 A0A224XXA4 A0A0A9WFR9 A0A0K8T9W4 A0A336M2S7 A0A2S2QL02 J9LG32
Ontologies
GO
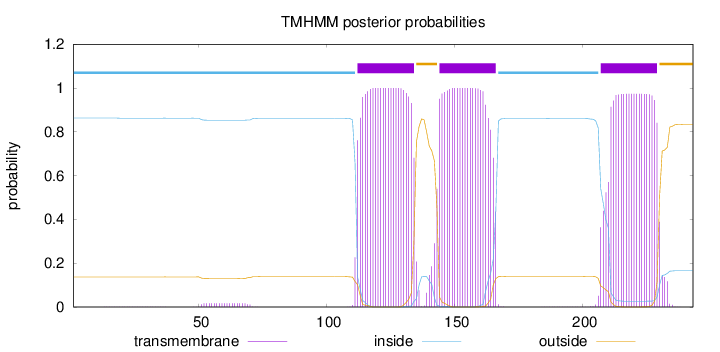
Topology
Length:
243
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
66.78578
Exp number, first 60 AAs:
0.19029
Total prob of N-in:
0.86276
inside
1 - 111
TMhelix
112 - 134
outside
135 - 143
TMhelix
144 - 166
inside
167 - 206
TMhelix
207 - 229
outside
230 - 243
Population Genetic Test Statistics
Pi
181.831033
Theta
148.320149
Tajima's D
0.310254
CLR
1.172939
CSRT
0.458627068646568
Interpretation
Uncertain
