Pre Gene Modal
BGIBMGA007000
Annotation
Protein_HGV2_[Papilio_machaon]
Location in the cell
Nuclear Reliability : 2.427
Sequence
CDS
ATGGCTGATATGATTGAAGTATCAACAGCAACTCCGGCGGAGTTACTTGGTGCTGGTCGACGCCACTTGGCTGTTAGAGACTTTCATTCGGCTGCTGAAGTACTGGCCAAAGCTTGTGAACTGTTGGCTAAAGAACATGGTGACACGGCGGACCAGTGTGCTGAGGCATATTTATGGTATGGCCAAGCACTATTAGGTTTGTCCAGGGATGAAAGTGGAGTTTTGGGAGATGGTATGCCAGGATCAGGCAATGTTGATGATGACGATGAAGAACAAGATGATGATGACAACAATGACAATGAACAAACAGAACAAAGCTGCGAACCTGAAAAGAGTAAAACTGAACACAACGAGGCTGAATCTACTTCGATGGAGGCTGATACATCAACAAAGGAAGCTGAGTCGAAACCGGCACAATCTACAACTAGTGATGCACCGGGTGCAAGTTCAGAGGTCACTGATAATGCTGAACCTAGCACAAGCAATGATGATGCTAATGATGAATCTGTCGTAAATATGGATAGTGAAGATGATGTCGACAATTTACAGCTTGCTTGGGAAATGCTTGACCTGGCCCGCAGTATACTGTCGCGGCGCGCGGCGGGCGGGGGCGAGGGCGCGGGGGGCGGCGCGGCGGCGCGGGCGCTGCTCGCCGACACGCACCTCGCGCTCGGGGAGGTCGCCCTCGAGAGCGAAACGTACGATAAAGCTGTTCTTGATATGCAGAGCTGCTTGGAGATCCAGAAGGAATTGTATCACAGCGACGACCGGCGTATTGCTGAAACACATTATCAAATTGGGCTGGCTAATTCCCTCGCTTCAAACTTCGAAGACGCGGTCACTCACTTCAAGAACGCCGCCAATATACTCGAAACCAGAATAAAGACTCTCGAGGGTCCAACCGCAGCCACGGACGACGCCACCGTCAAGAAGTACTCTTCAGCGGACCCGCTCTATACAATCGAAGGGGAGATCAAGGAATTGAAGGAACTTCTGCCGGAGATCCAAGAGAAAATACAAGACATGATGGATTATAAAGCGGAGACAATAAAACGGGTACGCGAAACCCTGTGCCCGTCGAACGGCGAGAGCTCTACACAAAATGGCGCTGGCTCTAGCGCGTCTCAGCCGCGACCGGCGGCTTCAGACATCTCGCATCTCATCAAGAGGAAACGGAAGGCGAGCAACGAACCCGAGGCCTCACCCGCGAAGAAGATGAACACGTGA
Protein
MADMIEVSTATPAELLGAGRRHLAVRDFHSAAEVLAKACELLAKEHGDTADQCAEAYLWYGQALLGLSRDESGVLGDGMPGSGNVDDDDEEQDDDDNNDNEQTEQSCEPEKSKTEHNEAESTSMEADTSTKEAESKPAQSTTSDAPGASSEVTDNAEPSTSNDDANDESVVNMDSEDDVDNLQLAWEMLDLARSILSRRAAGGGEGAGGGAAARALLADTHLALGEVALESETYDKAVLDMQSCLEIQKELYHSDDRRIAETHYQIGLANSLASNFEDAVTHFKNAANILETRIKTLEGPTAATDDATVKKYSSADPLYTIEGEIKELKELLPEIQEKIQDMMDYKAETIKRVRETLCPSNGESSTQNGAGSSASQPRPAASDISHLIKRKRKASNEPEASPAKKMNT
Summary
Uniprot
H9JBV5
A0A2A4ITI6
A0A2H1VJI6
A0A3S2PHI2
A0A194QZ08
A0A194PK27
+ More
A0A212EXZ8 S4PBX8 A0A1B6EV64 A0A195CRR4 E2C4L8 A0A1B6IXI9 A0A0J7KTG3 E2AB21 A0A067QPJ4 A0A1B6DE97 K7IY03 A0A2J7Q594 A0A2J7Q5A3 A0A2P8Z7C4 A0A1Z5LBI0 T1J7G1 A0A0C9RKU6 A0A2J7Q5A1 A0A293MXL7 C0HD91 A0A1S3LZP0 L7MJZ5 A0A131XAE3 A0A224YJK1 A0A1U7UNY9 A0A3B4U0G3 A0A060XAN2 A0A3Q0GSJ2 K9IXV7 A0A2Y9IU37 A0A341BGJ2 A0A3Q0CV51 A0A3Q0SKA0 A0A088AR75 E0VU41 A0A224XI19 A0A2I2Z1E4 K7FW38 A0A1A8C2M4 G9KCI8 A0A2Y9QHM9 A0A3P8PP92 A0A384CRP6 A0A3Q7MXY4 A0A384CRR0 A0A3Q7WID7 F1PLT4 A0A146VEQ0 A0A2Y9GRK2 A0A2K5E4R0 A0A3B4GWK8 A0A1V9XPK5 A0A1Y1MUX3 A0A250Y127 A0A2K5E4W1 A5D969 A0A2Y9GL39 T1I7P5 F6QS28 A0A287CWU6 A0A0V0G5K5 A0A2A3E9F0 A0A0L7REX9 A0A0P4VQQ3 A0A337SSX0 A0A2H8TMS4 A0A2H8TY52 A0A3P8ZG85 A0A1S3APW4 A0A3Q2VNG1 V4AN81 A0A3P8XER2 G3PUP7 A0A310SDK9 A0A1S3F8T4 A0A2R8N876 J9K8C9 A0A1W5A0J1 A0A023EYY4 A0A1S8WGZ4 A0A3B3BUN6 A0A183RP01 A0A075A523 A0A3B3BTV3
A0A212EXZ8 S4PBX8 A0A1B6EV64 A0A195CRR4 E2C4L8 A0A1B6IXI9 A0A0J7KTG3 E2AB21 A0A067QPJ4 A0A1B6DE97 K7IY03 A0A2J7Q594 A0A2J7Q5A3 A0A2P8Z7C4 A0A1Z5LBI0 T1J7G1 A0A0C9RKU6 A0A2J7Q5A1 A0A293MXL7 C0HD91 A0A1S3LZP0 L7MJZ5 A0A131XAE3 A0A224YJK1 A0A1U7UNY9 A0A3B4U0G3 A0A060XAN2 A0A3Q0GSJ2 K9IXV7 A0A2Y9IU37 A0A341BGJ2 A0A3Q0CV51 A0A3Q0SKA0 A0A088AR75 E0VU41 A0A224XI19 A0A2I2Z1E4 K7FW38 A0A1A8C2M4 G9KCI8 A0A2Y9QHM9 A0A3P8PP92 A0A384CRP6 A0A3Q7MXY4 A0A384CRR0 A0A3Q7WID7 F1PLT4 A0A146VEQ0 A0A2Y9GRK2 A0A2K5E4R0 A0A3B4GWK8 A0A1V9XPK5 A0A1Y1MUX3 A0A250Y127 A0A2K5E4W1 A5D969 A0A2Y9GL39 T1I7P5 F6QS28 A0A287CWU6 A0A0V0G5K5 A0A2A3E9F0 A0A0L7REX9 A0A0P4VQQ3 A0A337SSX0 A0A2H8TMS4 A0A2H8TY52 A0A3P8ZG85 A0A1S3APW4 A0A3Q2VNG1 V4AN81 A0A3P8XER2 G3PUP7 A0A310SDK9 A0A1S3F8T4 A0A2R8N876 J9K8C9 A0A1W5A0J1 A0A023EYY4 A0A1S8WGZ4 A0A3B3BUN6 A0A183RP01 A0A075A523 A0A3B3BTV3
Pubmed
19121390
26354079
22118469
23622113
20798317
24845553
+ More
20075255 29403074 28528879 20433749 25576852 28049606 28797301 24755649 26319212 20566863 22398555 17381049 23236062 24813606 16341006 28327890 28004739 28087693 12140684 16305752 19393038 27129103 17975172 25069045 23254933 25474469 29451363
20075255 29403074 28528879 20433749 25576852 28049606 28797301 24755649 26319212 20566863 22398555 17381049 23236062 24813606 16341006 28327890 28004739 28087693 12140684 16305752 19393038 27129103 17975172 25069045 23254933 25474469 29451363
EMBL
BABH01014929
NWSH01008657
PCG62460.1
ODYU01002912
SOQ40999.1
RSAL01000031
+ More
RVE51622.1 KQ461137 KPJ08831.1 KQ459603 KPI93079.1 AGBW02011637 OWR46366.1 GAIX01004401 JAA88159.1 GECZ01027932 JAS41837.1 KQ977349 KYN03396.1 GL452536 EFN77098.1 GECU01016083 JAS91623.1 LBMM01003369 KMQ93603.1 GL438234 EFN69297.1 KK853162 KDR10439.1 GEDC01013277 JAS24021.1 NEVH01017587 PNF23754.1 PNF23755.1 PYGN01000163 PSN52394.1 GFJQ02002392 JAW04578.1 JH431929 GBYB01007144 GBYB01013932 JAG76911.1 JAG83699.1 PNF23756.1 GFWV01019372 MAA44100.1 BT060297 ACN12657.1 GACK01001530 JAA63504.1 GEFH01003967 JAP64614.1 GFPF01006660 MAA17806.1 FR904936 CDQ73915.1 GABZ01006059 JAA47466.1 DS235779 EEB16897.1 GFTR01008336 JAW08090.1 CABD030002915 CABD030002916 CABD030002917 CABD030002918 AGCU01047627 AGCU01047628 HADZ01009392 SBP73333.1 JP014015 AES02613.1 AAEX03009779 AAEX03009780 GCES01070886 JAR15437.1 MNPL01006362 OQR75440.1 GEZM01026174 JAV87177.1 GFFV01002652 JAV37293.1 BT030488 ABQ12928.1 ACPB03000176 AGTP01008745 AGTP01008746 AGTP01008747 AGTP01008748 GECL01002838 JAP03286.1 KZ288313 PBC28357.1 KQ414608 KOC69369.1 GDKW01001777 JAI54818.1 AANG04003279 GFXV01003475 MBW15280.1 GFXV01006273 MBW18078.1 KB199650 ESP05629.1 KQ767621 OAD53383.1 ABLF02018459 GBBI01004823 JAC13889.1 KV907170 OON13722.1 KL596893 KER22529.1
RVE51622.1 KQ461137 KPJ08831.1 KQ459603 KPI93079.1 AGBW02011637 OWR46366.1 GAIX01004401 JAA88159.1 GECZ01027932 JAS41837.1 KQ977349 KYN03396.1 GL452536 EFN77098.1 GECU01016083 JAS91623.1 LBMM01003369 KMQ93603.1 GL438234 EFN69297.1 KK853162 KDR10439.1 GEDC01013277 JAS24021.1 NEVH01017587 PNF23754.1 PNF23755.1 PYGN01000163 PSN52394.1 GFJQ02002392 JAW04578.1 JH431929 GBYB01007144 GBYB01013932 JAG76911.1 JAG83699.1 PNF23756.1 GFWV01019372 MAA44100.1 BT060297 ACN12657.1 GACK01001530 JAA63504.1 GEFH01003967 JAP64614.1 GFPF01006660 MAA17806.1 FR904936 CDQ73915.1 GABZ01006059 JAA47466.1 DS235779 EEB16897.1 GFTR01008336 JAW08090.1 CABD030002915 CABD030002916 CABD030002917 CABD030002918 AGCU01047627 AGCU01047628 HADZ01009392 SBP73333.1 JP014015 AES02613.1 AAEX03009779 AAEX03009780 GCES01070886 JAR15437.1 MNPL01006362 OQR75440.1 GEZM01026174 JAV87177.1 GFFV01002652 JAV37293.1 BT030488 ABQ12928.1 ACPB03000176 AGTP01008745 AGTP01008746 AGTP01008747 AGTP01008748 GECL01002838 JAP03286.1 KZ288313 PBC28357.1 KQ414608 KOC69369.1 GDKW01001777 JAI54818.1 AANG04003279 GFXV01003475 MBW15280.1 GFXV01006273 MBW18078.1 KB199650 ESP05629.1 KQ767621 OAD53383.1 ABLF02018459 GBBI01004823 JAC13889.1 KV907170 OON13722.1 KL596893 KER22529.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000053268
UP000007151
+ More
UP000078542 UP000008237 UP000036403 UP000000311 UP000027135 UP000002358 UP000235965 UP000245037 UP000087266 UP000189704 UP000261420 UP000193380 UP000189705 UP000248482 UP000252040 UP000189706 UP000261340 UP000005203 UP000009046 UP000001519 UP000007267 UP000248480 UP000265100 UP000261680 UP000286641 UP000291021 UP000286642 UP000002254 UP000248481 UP000233020 UP000261460 UP000192247 UP000015103 UP000009136 UP000005215 UP000242457 UP000053825 UP000011712 UP000265140 UP000079721 UP000264840 UP000030746 UP000007635 UP000081671 UP000008225 UP000007819 UP000192224 UP000261560 UP000050792
UP000078542 UP000008237 UP000036403 UP000000311 UP000027135 UP000002358 UP000235965 UP000245037 UP000087266 UP000189704 UP000261420 UP000193380 UP000189705 UP000248482 UP000252040 UP000189706 UP000261340 UP000005203 UP000009046 UP000001519 UP000007267 UP000248480 UP000265100 UP000261680 UP000286641 UP000291021 UP000286642 UP000002254 UP000248481 UP000233020 UP000261460 UP000192247 UP000015103 UP000009136 UP000005215 UP000242457 UP000053825 UP000011712 UP000265140 UP000079721 UP000264840 UP000030746 UP000007635 UP000081671 UP000008225 UP000007819 UP000192224 UP000261560 UP000050792
Interpro
SUPFAM
SSF48452
SSF48452
Gene 3D
ProteinModelPortal
H9JBV5
A0A2A4ITI6
A0A2H1VJI6
A0A3S2PHI2
A0A194QZ08
A0A194PK27
+ More
A0A212EXZ8 S4PBX8 A0A1B6EV64 A0A195CRR4 E2C4L8 A0A1B6IXI9 A0A0J7KTG3 E2AB21 A0A067QPJ4 A0A1B6DE97 K7IY03 A0A2J7Q594 A0A2J7Q5A3 A0A2P8Z7C4 A0A1Z5LBI0 T1J7G1 A0A0C9RKU6 A0A2J7Q5A1 A0A293MXL7 C0HD91 A0A1S3LZP0 L7MJZ5 A0A131XAE3 A0A224YJK1 A0A1U7UNY9 A0A3B4U0G3 A0A060XAN2 A0A3Q0GSJ2 K9IXV7 A0A2Y9IU37 A0A341BGJ2 A0A3Q0CV51 A0A3Q0SKA0 A0A088AR75 E0VU41 A0A224XI19 A0A2I2Z1E4 K7FW38 A0A1A8C2M4 G9KCI8 A0A2Y9QHM9 A0A3P8PP92 A0A384CRP6 A0A3Q7MXY4 A0A384CRR0 A0A3Q7WID7 F1PLT4 A0A146VEQ0 A0A2Y9GRK2 A0A2K5E4R0 A0A3B4GWK8 A0A1V9XPK5 A0A1Y1MUX3 A0A250Y127 A0A2K5E4W1 A5D969 A0A2Y9GL39 T1I7P5 F6QS28 A0A287CWU6 A0A0V0G5K5 A0A2A3E9F0 A0A0L7REX9 A0A0P4VQQ3 A0A337SSX0 A0A2H8TMS4 A0A2H8TY52 A0A3P8ZG85 A0A1S3APW4 A0A3Q2VNG1 V4AN81 A0A3P8XER2 G3PUP7 A0A310SDK9 A0A1S3F8T4 A0A2R8N876 J9K8C9 A0A1W5A0J1 A0A023EYY4 A0A1S8WGZ4 A0A3B3BUN6 A0A183RP01 A0A075A523 A0A3B3BTV3
A0A212EXZ8 S4PBX8 A0A1B6EV64 A0A195CRR4 E2C4L8 A0A1B6IXI9 A0A0J7KTG3 E2AB21 A0A067QPJ4 A0A1B6DE97 K7IY03 A0A2J7Q594 A0A2J7Q5A3 A0A2P8Z7C4 A0A1Z5LBI0 T1J7G1 A0A0C9RKU6 A0A2J7Q5A1 A0A293MXL7 C0HD91 A0A1S3LZP0 L7MJZ5 A0A131XAE3 A0A224YJK1 A0A1U7UNY9 A0A3B4U0G3 A0A060XAN2 A0A3Q0GSJ2 K9IXV7 A0A2Y9IU37 A0A341BGJ2 A0A3Q0CV51 A0A3Q0SKA0 A0A088AR75 E0VU41 A0A224XI19 A0A2I2Z1E4 K7FW38 A0A1A8C2M4 G9KCI8 A0A2Y9QHM9 A0A3P8PP92 A0A384CRP6 A0A3Q7MXY4 A0A384CRR0 A0A3Q7WID7 F1PLT4 A0A146VEQ0 A0A2Y9GRK2 A0A2K5E4R0 A0A3B4GWK8 A0A1V9XPK5 A0A1Y1MUX3 A0A250Y127 A0A2K5E4W1 A5D969 A0A2Y9GL39 T1I7P5 F6QS28 A0A287CWU6 A0A0V0G5K5 A0A2A3E9F0 A0A0L7REX9 A0A0P4VQQ3 A0A337SSX0 A0A2H8TMS4 A0A2H8TY52 A0A3P8ZG85 A0A1S3APW4 A0A3Q2VNG1 V4AN81 A0A3P8XER2 G3PUP7 A0A310SDK9 A0A1S3F8T4 A0A2R8N876 J9K8C9 A0A1W5A0J1 A0A023EYY4 A0A1S8WGZ4 A0A3B3BUN6 A0A183RP01 A0A075A523 A0A3B3BTV3
Ontologies
KEGG
GO
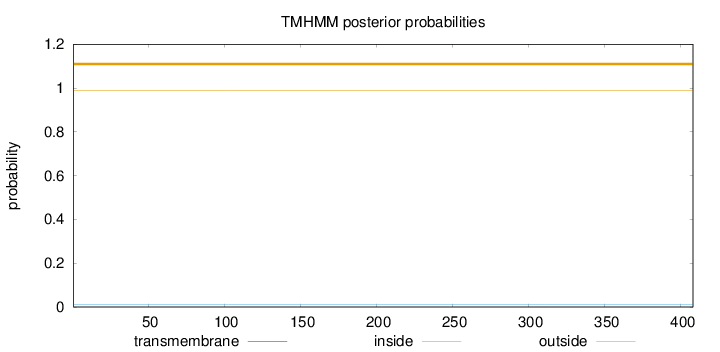
Topology
Length:
408
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00063
Exp number, first 60 AAs:
0.00014
Total prob of N-in:
0.01236
outside
1 - 408
Population Genetic Test Statistics
Pi
194.14402
Theta
153.063191
Tajima's D
0.961154
CLR
0.575571
CSRT
0.650367481625919
Interpretation
Uncertain
