Gene
KWMTBOMO10464 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007098
Annotation
NADP-dependent_oxidoreductase_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 3.079
Sequence
CDS
ATGACCTCAGCTAGGAAGTACGTCGTGAAGAACCACTTTAAAGGTGTACCAAAACAAGAGGACTACGAATTAGTCGAATTTGTTATACCGCCTCTGATCGACGGCGAGGTCCTCGTTAAAGCCGAATGGATCAGCGTCGACCCGTATTTAAGGGCTTACAACTCTTACCAAGCTGTACCCTACGACCAGTTCAGTTATCAAGTCGGTGTTGTTGTCCAATCAAGGGATAGCAACTATCCGGTAGGAACGAGAGTTGTTGCGCACAAAGGATGGTGCGACCACTACGTCTTCACTCCTAGTACGCAACCTAACACTCCAAAGGATCGTATTTATAAACTACCAGATCTGCAGGGATTGTCGCCGTCGCTGGGCGTAGGGGCGGTTGGGATGCCGGGCGCGACTGCCTATTTTGGTTTCTTAGAAATCTGCAAGCCAAAAGCTGGCGAGACCGTGGTGGTCACCGGAGCGGCCGGTGCTGTGGGATCACTTGTCGGTCAAATAGCTAAGATCAAGGGCTGCAGAGTCATCGGCTTCGCTGGCACGGATGATAAAGTGAAATGGCTGGAGGAAGAGCTCGGCTTCGACAAGGCGTTTAATTATAAAACAGTAGATGTACCGGCAGCACTCAAGGAAGCTGCCCCTAACGGGATAGACTGTTATTTCGACAATGTCGGCGGTGAAATCAGCAGTCAGATAATTAGCAAAATGAACGTTTACGGGAGGGTCTCCGTCTGTGGCAGCATAAGCGCGTACAATGAGGATCTAACTAAATTACCCAAAGCGACTATTCTGCAACCTAGTTTAGTATTCAACCAAATCAAAGTCGAAGGTTTTCTAGTTTGGCGCTGGAATGATCAGTCTAAGGCGTTTGCGGAAATCATTCCTTGGATTCAGAGTGGGAAATTGAAAGTAAAGGAACATGTCACGGAAGGATTCGATAAACTCTTTGATGCCTTTATAGGAATGCTTGCCGGGGAGAATTACGGAAAGGCCGTAGTCAAAGTGTAA
Protein
MTSARKYVVKNHFKGVPKQEDYELVEFVIPPLIDGEVLVKAEWISVDPYLRAYNSYQAVPYDQFSYQVGVVVQSRDSNYPVGTRVVAHKGWCDHYVFTPSTQPNTPKDRIYKLPDLQGLSPSLGVGAVGMPGATAYFGFLEICKPKAGETVVVTGAAGAVGSLVGQIAKIKGCRVIGFAGTDDKVKWLEEELGFDKAFNYKTVDVPAALKEAAPNGIDCYFDNVGGEISSQIISKMNVYGRVSVCGSISAYNEDLTKLPKATILQPSLVFNQIKVEGFLVWRWNDQSKAFAEIIPWIQSGKLKVKEHVTEGFDKLFDAFIGMLAGENYGKAVVKV
Summary
Uniprot
Q1HPR4
A0A194QU38
A0A194PI33
I4DRQ9
A0A0H4TDB4
H9JC55
+ More
A0A1L8D6Z1 A0A1L8D6M5 A0A0L7K5Q5 A0A0L7L3S9 A0A212EY34 A0A194PID5 A0A212EJX1 A0A2A4J3P3 A0A2A4J403 U5KCL5 A0A0F6Q0V0 A0A194QV25 A0A0F6Q1F9 A0A194PJ62 A0A2A4J243 A0A2A4J3L6 A0A2H1VJJ7 A0A2A4J210 A0A232EYT1 K7IZQ2 A0A2A4J401 D2SNL0 A0A0C9QI54 A0A1B6H016 A0A151XA86 A0A194PWW2 A0A151XA81 A0A151ITC7 A0A195AV65 A0A076FRA6 A0A194QU86 A0A0P5Y8F0 A0A0P5F538 A0A158NN98 A0A0F7QHK0 A0A0L7QX07 A0A212FPV5 A0A195FY34 A0A0P4WLV0 A0A0P5S5A7 A0A195CRZ6 A0A0P5SYK7 A0A0P5RGK0 A0A0P5LKL0 A0A0F6Q1G9 A0A0P5X055 A0A1W4WHR3 A0A0P5BM78 E9H9N6 A0A0P5EUQ4 A0A0P5GWK1 A0A0H4TG53 E9FXX5 A0A067RU96 A0A0P5NY48 A0A2J7RMM5 A0A0P4WLU2 A0A0N8A436 A0A2A3EK12 K7J6V5 A0A088AD26 A0A0P5Z4Y7 A0A0P5UYR3 A0A0K8TRN6 A0A067RFV9 A0A1J1I2L4 A0A0P5Y6A2 F4WLJ0 J3JTH7 A0A2S2R5N0 A0A195CRA2 A0A336MUL9 A0A0P5WI19 A0A2A4JNH3 Q967C7 A0A0P5SWF3 A0A0P4XMF8 A0A026W461 J3JU26 A0A0P6GVR3 N6U1X5 A0A0P6AL25 A0A195E3V0 A0A3S2PGN0 F4WLJ1 A0A2J7QDG3 A0A0N0BDE4 A0A2J7QDH2 A0A195FVS8 A0A0P5HF88 K7J6V7 A0A2H1VZJ8 A0A0C9RL60 A0A0P5GMF1
A0A1L8D6Z1 A0A1L8D6M5 A0A0L7K5Q5 A0A0L7L3S9 A0A212EY34 A0A194PID5 A0A212EJX1 A0A2A4J3P3 A0A2A4J403 U5KCL5 A0A0F6Q0V0 A0A194QV25 A0A0F6Q1F9 A0A194PJ62 A0A2A4J243 A0A2A4J3L6 A0A2H1VJJ7 A0A2A4J210 A0A232EYT1 K7IZQ2 A0A2A4J401 D2SNL0 A0A0C9QI54 A0A1B6H016 A0A151XA86 A0A194PWW2 A0A151XA81 A0A151ITC7 A0A195AV65 A0A076FRA6 A0A194QU86 A0A0P5Y8F0 A0A0P5F538 A0A158NN98 A0A0F7QHK0 A0A0L7QX07 A0A212FPV5 A0A195FY34 A0A0P4WLV0 A0A0P5S5A7 A0A195CRZ6 A0A0P5SYK7 A0A0P5RGK0 A0A0P5LKL0 A0A0F6Q1G9 A0A0P5X055 A0A1W4WHR3 A0A0P5BM78 E9H9N6 A0A0P5EUQ4 A0A0P5GWK1 A0A0H4TG53 E9FXX5 A0A067RU96 A0A0P5NY48 A0A2J7RMM5 A0A0P4WLU2 A0A0N8A436 A0A2A3EK12 K7J6V5 A0A088AD26 A0A0P5Z4Y7 A0A0P5UYR3 A0A0K8TRN6 A0A067RFV9 A0A1J1I2L4 A0A0P5Y6A2 F4WLJ0 J3JTH7 A0A2S2R5N0 A0A195CRA2 A0A336MUL9 A0A0P5WI19 A0A2A4JNH3 Q967C7 A0A0P5SWF3 A0A0P4XMF8 A0A026W461 J3JU26 A0A0P6GVR3 N6U1X5 A0A0P6AL25 A0A195E3V0 A0A3S2PGN0 F4WLJ1 A0A2J7QDG3 A0A0N0BDE4 A0A2J7QDH2 A0A195FVS8 A0A0P5HF88 K7J6V7 A0A2H1VZJ8 A0A0C9RL60 A0A0P5GMF1
Pubmed
EMBL
BABH01014929
DQ443338
ABF51427.1
KQ461137
KPJ08829.1
KQ459603
+ More
KPI93076.1 AK405232 BAM20599.1 KP899554 AKQ06153.1 GEYN01000025 JAV02104.1 GEYN01000023 JAV02106.1 JTDY01010417 KOB58214.1 JTDY01003120 KOB70117.1 AGBW02011637 OWR46367.1 KPI93077.1 AGBW02014376 OWR41771.1 NWSH01003541 PCG66134.1 PCG66132.1 KC007342 AGQ45607.1 KP237874 AKD01727.1 KPJ08830.1 KP237886 AKD01739.1 KPI93078.1 PCG66135.1 NWSH01003564 PCG66104.1 ODYU01002912 SOQ40998.1 PCG66105.1 NNAY01001646 OXU23328.1 PCG66133.1 EZ407138 ACX53701.1 GBYB01000237 JAG70004.1 GECZ01021917 GECZ01001753 GECZ01001434 JAS47852.1 JAS68016.1 JAS68335.1 KQ982351 KYQ57274.1 KQ459595 KPI95625.1 KYQ57275.1 KQ981037 KYN10151.1 KQ976738 KYM75864.1 KF960800 AII22002.1 KQ461154 KPJ08550.1 GDIP01220647 GDIP01062315 JAM41400.1 GDIQ01265642 JAJ86082.1 ADTU01021185 ADTU01021186 LC017750 BAR64764.1 KQ414706 KOC63074.1 AGBW02000285 OWR55798.1 KQ981208 KYN44744.1 GDIP01255092 JAI68309.1 GDIQ01096505 GDIP01111355 GDIP01043199 GDIQ01054177 LRGB01000190 JAL55221.1 JAL92359.1 KZS20464.1 KQ977372 KYN03267.1 GDIQ01187424 GDIP01140398 JAK64301.1 JAL63316.1 GDIQ01110399 JAL41327.1 GDIQ01170510 JAK81215.1 KP237896 AKD01749.1 GDIP01079654 JAM24061.1 GDIP01183042 JAJ40360.1 GL732609 EFX71508.1 GDIQ01269885 JAJ81839.1 GDIQ01236708 JAK15017.1 KP899553 AKQ06152.1 GL732526 EFX88216.1 KK852415 KDR24385.1 GDIQ01138237 JAL13489.1 NEVH01002552 PNF42096.1 GDIP01253621 JAI69780.1 GDIP01184449 JAJ38953.1 KZ288220 PBC32143.1 AAZX01013836 GDIP01049065 JAM54650.1 GDIP01109651 JAL94063.1 GDAI01001023 JAI16580.1 KK852493 KDR22751.1 CVRI01000036 CRK93094.1 GDIP01062314 JAM41401.1 GL888208 EGI64926.1 BT126534 AEE61498.1 GGMS01016030 MBY85233.1 KYN03268.1 UFQS01002035 UFQT01002035 SSX13033.1 SSX32473.1 GDIP01086512 JAM17203.1 NWSH01000966 PCG73369.1 Y19102 CAC38761.1 GDIP01137302 JAL66412.1 GDIP01240844 LRGB01001361 JAI82557.1 KZS12747.1 KK107446 EZA50798.1 BT126738 AEE61700.1 GDIQ01028521 JAN66216.1 APGK01045490 KB741037 KB632085 ENN74606.1 ERL88639.1 GDIP01027804 JAM75911.1 KQ979685 KYN19823.1 RSAL01000041 RVE50877.1 EGI64927.1 NEVH01015823 PNF26620.1 KQ435878 KOX69991.1 PNF26623.1 KYN44745.1 GDIQ01250759 JAK00966.1 AAZX01001296 ODYU01005116 SOQ45664.1 GBYB01007656 JAG77423.1 GDIQ01239153 JAK12572.1
KPI93076.1 AK405232 BAM20599.1 KP899554 AKQ06153.1 GEYN01000025 JAV02104.1 GEYN01000023 JAV02106.1 JTDY01010417 KOB58214.1 JTDY01003120 KOB70117.1 AGBW02011637 OWR46367.1 KPI93077.1 AGBW02014376 OWR41771.1 NWSH01003541 PCG66134.1 PCG66132.1 KC007342 AGQ45607.1 KP237874 AKD01727.1 KPJ08830.1 KP237886 AKD01739.1 KPI93078.1 PCG66135.1 NWSH01003564 PCG66104.1 ODYU01002912 SOQ40998.1 PCG66105.1 NNAY01001646 OXU23328.1 PCG66133.1 EZ407138 ACX53701.1 GBYB01000237 JAG70004.1 GECZ01021917 GECZ01001753 GECZ01001434 JAS47852.1 JAS68016.1 JAS68335.1 KQ982351 KYQ57274.1 KQ459595 KPI95625.1 KYQ57275.1 KQ981037 KYN10151.1 KQ976738 KYM75864.1 KF960800 AII22002.1 KQ461154 KPJ08550.1 GDIP01220647 GDIP01062315 JAM41400.1 GDIQ01265642 JAJ86082.1 ADTU01021185 ADTU01021186 LC017750 BAR64764.1 KQ414706 KOC63074.1 AGBW02000285 OWR55798.1 KQ981208 KYN44744.1 GDIP01255092 JAI68309.1 GDIQ01096505 GDIP01111355 GDIP01043199 GDIQ01054177 LRGB01000190 JAL55221.1 JAL92359.1 KZS20464.1 KQ977372 KYN03267.1 GDIQ01187424 GDIP01140398 JAK64301.1 JAL63316.1 GDIQ01110399 JAL41327.1 GDIQ01170510 JAK81215.1 KP237896 AKD01749.1 GDIP01079654 JAM24061.1 GDIP01183042 JAJ40360.1 GL732609 EFX71508.1 GDIQ01269885 JAJ81839.1 GDIQ01236708 JAK15017.1 KP899553 AKQ06152.1 GL732526 EFX88216.1 KK852415 KDR24385.1 GDIQ01138237 JAL13489.1 NEVH01002552 PNF42096.1 GDIP01253621 JAI69780.1 GDIP01184449 JAJ38953.1 KZ288220 PBC32143.1 AAZX01013836 GDIP01049065 JAM54650.1 GDIP01109651 JAL94063.1 GDAI01001023 JAI16580.1 KK852493 KDR22751.1 CVRI01000036 CRK93094.1 GDIP01062314 JAM41401.1 GL888208 EGI64926.1 BT126534 AEE61498.1 GGMS01016030 MBY85233.1 KYN03268.1 UFQS01002035 UFQT01002035 SSX13033.1 SSX32473.1 GDIP01086512 JAM17203.1 NWSH01000966 PCG73369.1 Y19102 CAC38761.1 GDIP01137302 JAL66412.1 GDIP01240844 LRGB01001361 JAI82557.1 KZS12747.1 KK107446 EZA50798.1 BT126738 AEE61700.1 GDIQ01028521 JAN66216.1 APGK01045490 KB741037 KB632085 ENN74606.1 ERL88639.1 GDIP01027804 JAM75911.1 KQ979685 KYN19823.1 RSAL01000041 RVE50877.1 EGI64927.1 NEVH01015823 PNF26620.1 KQ435878 KOX69991.1 PNF26623.1 KYN44745.1 GDIQ01250759 JAK00966.1 AAZX01001296 ODYU01005116 SOQ45664.1 GBYB01007656 JAG77423.1 GDIQ01239153 JAK12572.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000037510
UP000007151
UP000218220
+ More
UP000215335 UP000002358 UP000075809 UP000078492 UP000078540 UP000005205 UP000053825 UP000078541 UP000076858 UP000078542 UP000192223 UP000000305 UP000027135 UP000235965 UP000242457 UP000005203 UP000183832 UP000007755 UP000053097 UP000019118 UP000030742 UP000283053 UP000053105
UP000215335 UP000002358 UP000075809 UP000078492 UP000078540 UP000005205 UP000053825 UP000078541 UP000076858 UP000078542 UP000192223 UP000000305 UP000027135 UP000235965 UP000242457 UP000005203 UP000183832 UP000007755 UP000053097 UP000019118 UP000030742 UP000283053 UP000053105
PRIDE
Interpro
ProteinModelPortal
Q1HPR4
A0A194QU38
A0A194PI33
I4DRQ9
A0A0H4TDB4
H9JC55
+ More
A0A1L8D6Z1 A0A1L8D6M5 A0A0L7K5Q5 A0A0L7L3S9 A0A212EY34 A0A194PID5 A0A212EJX1 A0A2A4J3P3 A0A2A4J403 U5KCL5 A0A0F6Q0V0 A0A194QV25 A0A0F6Q1F9 A0A194PJ62 A0A2A4J243 A0A2A4J3L6 A0A2H1VJJ7 A0A2A4J210 A0A232EYT1 K7IZQ2 A0A2A4J401 D2SNL0 A0A0C9QI54 A0A1B6H016 A0A151XA86 A0A194PWW2 A0A151XA81 A0A151ITC7 A0A195AV65 A0A076FRA6 A0A194QU86 A0A0P5Y8F0 A0A0P5F538 A0A158NN98 A0A0F7QHK0 A0A0L7QX07 A0A212FPV5 A0A195FY34 A0A0P4WLV0 A0A0P5S5A7 A0A195CRZ6 A0A0P5SYK7 A0A0P5RGK0 A0A0P5LKL0 A0A0F6Q1G9 A0A0P5X055 A0A1W4WHR3 A0A0P5BM78 E9H9N6 A0A0P5EUQ4 A0A0P5GWK1 A0A0H4TG53 E9FXX5 A0A067RU96 A0A0P5NY48 A0A2J7RMM5 A0A0P4WLU2 A0A0N8A436 A0A2A3EK12 K7J6V5 A0A088AD26 A0A0P5Z4Y7 A0A0P5UYR3 A0A0K8TRN6 A0A067RFV9 A0A1J1I2L4 A0A0P5Y6A2 F4WLJ0 J3JTH7 A0A2S2R5N0 A0A195CRA2 A0A336MUL9 A0A0P5WI19 A0A2A4JNH3 Q967C7 A0A0P5SWF3 A0A0P4XMF8 A0A026W461 J3JU26 A0A0P6GVR3 N6U1X5 A0A0P6AL25 A0A195E3V0 A0A3S2PGN0 F4WLJ1 A0A2J7QDG3 A0A0N0BDE4 A0A2J7QDH2 A0A195FVS8 A0A0P5HF88 K7J6V7 A0A2H1VZJ8 A0A0C9RL60 A0A0P5GMF1
A0A1L8D6Z1 A0A1L8D6M5 A0A0L7K5Q5 A0A0L7L3S9 A0A212EY34 A0A194PID5 A0A212EJX1 A0A2A4J3P3 A0A2A4J403 U5KCL5 A0A0F6Q0V0 A0A194QV25 A0A0F6Q1F9 A0A194PJ62 A0A2A4J243 A0A2A4J3L6 A0A2H1VJJ7 A0A2A4J210 A0A232EYT1 K7IZQ2 A0A2A4J401 D2SNL0 A0A0C9QI54 A0A1B6H016 A0A151XA86 A0A194PWW2 A0A151XA81 A0A151ITC7 A0A195AV65 A0A076FRA6 A0A194QU86 A0A0P5Y8F0 A0A0P5F538 A0A158NN98 A0A0F7QHK0 A0A0L7QX07 A0A212FPV5 A0A195FY34 A0A0P4WLV0 A0A0P5S5A7 A0A195CRZ6 A0A0P5SYK7 A0A0P5RGK0 A0A0P5LKL0 A0A0F6Q1G9 A0A0P5X055 A0A1W4WHR3 A0A0P5BM78 E9H9N6 A0A0P5EUQ4 A0A0P5GWK1 A0A0H4TG53 E9FXX5 A0A067RU96 A0A0P5NY48 A0A2J7RMM5 A0A0P4WLU2 A0A0N8A436 A0A2A3EK12 K7J6V5 A0A088AD26 A0A0P5Z4Y7 A0A0P5UYR3 A0A0K8TRN6 A0A067RFV9 A0A1J1I2L4 A0A0P5Y6A2 F4WLJ0 J3JTH7 A0A2S2R5N0 A0A195CRA2 A0A336MUL9 A0A0P5WI19 A0A2A4JNH3 Q967C7 A0A0P5SWF3 A0A0P4XMF8 A0A026W461 J3JU26 A0A0P6GVR3 N6U1X5 A0A0P6AL25 A0A195E3V0 A0A3S2PGN0 F4WLJ1 A0A2J7QDG3 A0A0N0BDE4 A0A2J7QDH2 A0A195FVS8 A0A0P5HF88 K7J6V7 A0A2H1VZJ8 A0A0C9RL60 A0A0P5GMF1
PDB
2DM6
E-value=4.11994e-74,
Score=706
Ontologies
GO
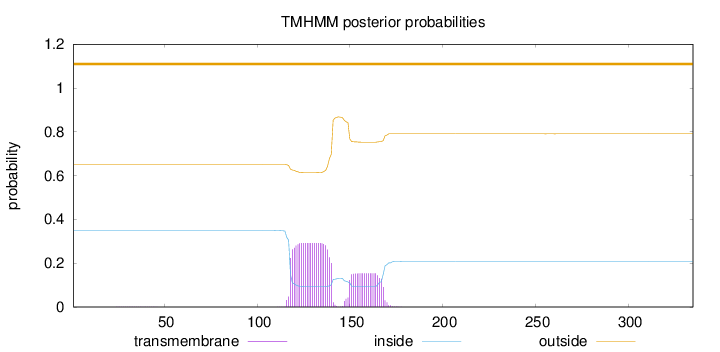
Topology
Length:
335
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
9.46018
Exp number, first 60 AAs:
0.00403
Total prob of N-in:
0.34929
outside
1 - 335
Population Genetic Test Statistics
Pi
142.56903
Theta
161.282745
Tajima's D
0.264341
CLR
0.397417
CSRT
0.44002799860007
Interpretation
Uncertain
