Gene
KWMTBOMO10456
Pre Gene Modal
BGIBMGA007095
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_protein_CLEC16A_[Bombyx_mori]
Full name
Protein CLEC16A homolog
Alternative Name
Endosomal maturation defective protein
Location in the cell
Nuclear Reliability : 1.804 PlasmaMembrane Reliability : 2.247
Sequence
CDS
ATGTTCCGGAGCAGAAGCTGGTTCGGGACTGGCTGGGGCCGTCCCAAGAACCCCCATTCATTGGAAAGACTAAAATATTTACACAATATACTTTGCAAAAATACAACTGTTTCTGAAAGCAACAGAGGAACGCTTGTGGAATCGCTAAGGTGCATAGCGGAGATACTCATATGGGGCGATCAGAATGACAGCTCAGTTTTTGATTTCTTCTTAGAGAAGAACATGTTGTCATATTTCTTGAAGATAATGAGACAGAAGTGTGGAGGCTCATCATATGTATGTGTGCAGCTTTTGCAAACCCTGAATATACTGTTTGAGAACATCAGGAATGAAACATCACTGTATTACCTCTTAAGCAATAACCACGTTAATTCTATAATAGTTCATAAGTTCGATTTTTCCGATGAAGAAGTGATGGCATATTACATATCATTCCTGAAGACATTAAGTTTAAAACTCAACAACCACACGATACATTTCTTTTACAATGAGCACACAAAGGATTTTCCACTTTACACCGAGGCAATAAAGTTCTTTAACCATTCAGAGTCGATGGTTCGGATTGCGGTGCGGACGCTCACCCTTAATGTGTATAGAGTACAGGATGCCAGCATGCTTAGATTTATAAGAGACAGGACGGCAGCGCCATATTTTAGTAATTTGGTCTGGTTTATCGGAAAACATATATTGGAGTTAGATACTTGTGTCAGGAACGACGCTGACCATCAATCCCAGCAAAAATTGGACGACTTAGTTGCCGAACATCTGGACCATCTACACTACATTAACGACATCCTATGCCTAAATATACCAGATCTTAACGACGTGCTCACGGAACATCTTCTACATAAACTGCTGATACCCTTATACATTTATTCTTTGACCTCTGAATCGATTAAAAGACCGTCTACGGCGCCCCCCTACAATAGGGATCATCTGCAAAACATAGAAGTTATAACAAGGAGCTTAGAGGCCATTTTGACGGGGAAAAACAGCGAAATGTCGCCTTCTAGAATGAACTTCCCGAGACAAAGAATCGAGAGCGAAGAAGAGAGACCTTCTGTATCGTGTGTGGTTGCCCTGTTTCTCTTATCGCAAGTGTTCCTAATAATAACACACGGCCCGATTGTGCACGCGTTGGCCTGGATCTTATTGCAATCAGACTTATCGGTGCTGGAAGAAGGAGCTACGAAAATTCTCGAAATGTACATCAGTACCGCTAAGCCCAATGTCAAATTGGAGTTTTCGAAGCCGCAGATGAGTCTAGAGAAGGCTTTAGAGAATAATTCTGGCGAACGCGATTACACTACACCGGAATACAGTGGCTGTAAAACGTTCAGTTTCGACGACAAAGATACAGATCATTCGAGTTGTGATCTCGATCCTGAGTTGGTATCGAATTCACTTCCGTCCACATCTCATATAAGCGAAACGTCGAGTATAGCTCACGACTCGACTGAAGATCTCGTGACTCAAATTCATTCGGTTAAATCGAGTGTCGTTCACAATGAGAACATGCCGGATTCTCCGCTGCCCGATTCTGGTATAGAATCCAGTTCTACGAAATCCAATTCGGAATTCAACAACATAACTGATGAAGAGAAACAGCAATTGCTCGGAGTCACAACTAGTACACCAATAAACAGATCCGATTCGCTAGAGAATGTATTGGAGAAAGAGAATCGAGAAATCGCCAAGAGGCCGTTTCTAAAGACGATCTTTGATTCTTTGGATTGTGGCGAGAATGATTATAAAGCATTATTTGCTCTTTGCTTATTGTTCGCGTTGATAAATAACAAAGGTGTCAACACAGAACTACTTGATACGCTGTTGTGTCCCGAAGAGGAACTGTCTCATTTGAACTCAGCGAATACGTCGGGCCGTGAAATTGTAGAGAAAAGCGAAGGATCATCGACATCAACTTCTTCTACTCCTACAAAAAAAACATTGCAGAGTTTTAACTCACTATTGATTTATAAGCTAATGGCTATTGCAAATTTGAGCTGTAAACCTACCTCTCAGGTCCGCCTGGTCACTCTGGAGTTGAGCGTGACGTTAATGCGCATCGCAATGCAAGGGACTAGGGGTCCCGCGCCGTCAGCTAACGGGAGCTCACGGTCAGCGACGGGCCACTCCGGCCCGGGCTCACGGAGCGTCATGAGCCGTCACGTGGCCGCCGCGGACACGTTACGTGCGCGGGCCGCAGCCCTCGCACGTAACTTTTACAAGTGCGACGATATCTTCCTGGACATGTTTGAAGATGAGTACTGTGAGATGAGCAAGCGTCCTCTGAACGTGGAATGGCTATGCATGGATGCGGCAATACTGCTTCCTCCCACACGCACGCCGATGTCTGGCATTGCGTTTGCTAAGCGACTGCCGAGCGGGGAGATAGAGAGAGCCCGTAGAGCGATAAGATCGTTCTTCCTAATACGAGACCTGTTCTTGAAGTTGAGCGGGATACCAGAAAACCAACTGCCTCTGACGAACCCGGCACCGTTTGTCGAAGTCGGCAACGTTTTGGATCTCAACGATTCCGATCTGATATCGTGCGACATAATACAGAAGGATGGTACGTGGCAGCACAGGTTCTTAGCCGTCGACAACATGCAGATAATACTCGTCGAGCCGGACAAACAGAGGCTCGGGTGGGGAGTCGCCAAACTGGTGGGCAGCCTTCAGGATTTGGAGATCCAAGGCGATAAGGAAGATCCGCGGTGCCTCCACCTGACCATCCACAAGCCGCGGGTGGGCGCGACGGGCGCGCTTCCCAGGACGCTACTGCTGCGGGCCAAGTTCAAGTTCGACGACTACATACGCAGCATGGCGGCCAAACAGAGGCTCATTAAGGGTCGCATGAAGTCCCGCCAGAAGAAGATGCAGCAGATCGGGCGACTGCTGGAAGTGTCGGGAGTGTCCGCGCCCCCCGCCGCGCCGCGCCCCCGCCCGCTGTTCTCCCGCCCCGGCCTCTCGGCGATCAGGAGATCTACAGGTTCCGTACAATTGCACGTTCTATCCAAACACAATTCGTCTTTTCTCATTGAAAGTGTACAATTTCCAAATTCGAAATTGAGTATTAACTCAATTTCGAATTTGGAACAGGAAAACTAA
Protein
MFRSRSWFGTGWGRPKNPHSLERLKYLHNILCKNTTVSESNRGTLVESLRCIAEILIWGDQNDSSVFDFFLEKNMLSYFLKIMRQKCGGSSYVCVQLLQTLNILFENIRNETSLYYLLSNNHVNSIIVHKFDFSDEEVMAYYISFLKTLSLKLNNHTIHFFYNEHTKDFPLYTEAIKFFNHSESMVRIAVRTLTLNVYRVQDASMLRFIRDRTAAPYFSNLVWFIGKHILELDTCVRNDADHQSQQKLDDLVAEHLDHLHYINDILCLNIPDLNDVLTEHLLHKLLIPLYIYSLTSESIKRPSTAPPYNRDHLQNIEVITRSLEAILTGKNSEMSPSRMNFPRQRIESEEERPSVSCVVALFLLSQVFLIITHGPIVHALAWILLQSDLSVLEEGATKILEMYISTAKPNVKLEFSKPQMSLEKALENNSGERDYTTPEYSGCKTFSFDDKDTDHSSCDLDPELVSNSLPSTSHISETSSIAHDSTEDLVTQIHSVKSSVVHNENMPDSPLPDSGIESSSTKSNSEFNNITDEEKQQLLGVTTSTPINRSDSLENVLEKENREIAKRPFLKTIFDSLDCGENDYKALFALCLLFALINNKGVNTELLDTLLCPEEELSHLNSANTSGREIVEKSEGSSTSTSSTPTKKTLQSFNSLLIYKLMAIANLSCKPTSQVRLVTLELSVTLMRIAMQGTRGPAPSANGSSRSATGHSGPGSRSVMSRHVAAADTLRARAAALARNFYKCDDIFLDMFEDEYCEMSKRPLNVEWLCMDAAILLPPTRTPMSGIAFAKRLPSGEIERARRAIRSFFLIRDLFLKLSGIPENQLPLTNPAPFVEVGNVLDLNDSDLISCDIIQKDGTWQHRFLAVDNMQIILVEPDKQRLGWGVAKLVGSLQDLEIQGDKEDPRCLHLTIHKPRVGATGALPRTLLLRAKFKFDDYIRSMAAKQRLIKGRMKSRQKKMQQIGRLLEVSGVSAPPAAPRPRPLFSRPGLSAIRRSTGSVQLHVLSKHNSSFLIESVQFPNSKLSINSISNLEQEN
Summary
Description
Required for mitophagy, autophagy and endosome maturation, possibly by acting in multiple membrane trafficking pathways (PubMed:20194640, PubMed:22493244). Required for endosome trafficking and maturation (PubMed:20194640). Functions with the class C Vps-HOPS complex member Vps16a to promote endosomal maturation into degradative late endosomes and lysosomes (PubMed:20194640). In response to starvation, functions at an early stage of autophagy to promote autophagosome growth and efficient autophagy (PubMed:22493244). Essential for the recruitment of lva-positive Golgi elements to autophagosomes (PubMed:22493244). Likely to function by promoting membrane traffic from the Golgi complex to the developing autophagosomes (PubMed:22493244). Also regulates synaptic growth at the neuromuscular junctions (NMJ) by down-regulating BMP signaling (PubMed:20194640).
Subunit
Interacts with the class C Vps-HOPS complex components; Car, Dor and Vps16a.
Similarity
Belongs to the CLEC16A/gop-1 family.
Keywords
Alternative splicing
Autophagy
Complete proteome
Cytoplasmic vesicle
Endosome
Golgi apparatus
Membrane
Reference proteome
Transmembrane
Transmembrane helix
Feature
chain Protein CLEC16A homolog
splice variant In isoform B.
splice variant In isoform B.
Uniprot
A0A3S2NMS2
A0A2H1VEA9
A0A212FDA9
A0A194QYZ8
A0A194PK18
A0A2A3E9D9
+ More
A0A088AR87 A0A0N0U4H2 E2BSP9 E2AWT9 A0A0C9Q4M4 F4W9K1 A0A158NJF5 A0A151WI43 A0A195DL19 A0A3L8DUS3 A0A232EEZ4 A0A2J7Q3N5 A0A0L0BQI8 A0A0A1WZ30 W8C1E8 A0A1I8P6P0 A0A0K8UEZ4 A0A1I8P6Q1 A0A034VR38 A0A0K8UGJ7 B3M2J8 A0A1I8ND12 A0A0P8Y0H1 B4NLA0 B4KBZ5 A0A182HTJ4 A0A195BHU1 A0A182XIS2 A0A182UNJ9 A0A195F3Y5 A0A0Q9X7Y1 A0A182LBX1 A0A182U6V7 B4JFL3 A0A067QNI2 A0A0M4EPJ2 A0A084W353 A0A0Q9X2R2 A0A1A9ZAC8 A0A1A9VD35 Q9VEV4-2 B4LX42 A0A0R1E2Z8 A0A1W4UIX9 A0A1B0G4T3 B4GFL5 Q9VEV4 A0A1W4UKR1 B4PQZ6 B3P123 A0A1B0AZE5 B4IBV9 Q295W4 A0A0R1E4F7 A0A0Q9WQ14 A0A1W4U778 A0A1W4U7U0 A0A0R1E3N5 A0A3B0KL53 I5APE5 B4QX33 T1J9I9 R7V952 A0A1S3HH86 A0A0L8I5W1 A0A0L8I6S0 A0A2T7PZ30 L7MC62 V5I041 A0A0P6J253 G5BWF2 A0A2Y9LLK4 A0A2Y9LEC0 A0A2Y9HQA9 A0A3Q7MVS5 A0A2Y9LGR7 A0A2U3W0C8 L9KXV9 A0A384AM87 A0A286Y0A2 A0A341BAZ0 A0A3Q7MTS3 A0A286XCM5 A0A3G9GRT0 F6TYT8 A0A3Q7P6M2 A0A2Y9EFK1 A0A2Y9EG90 F1MQK7 A0A384AM70 A0A3Q2HSZ4
A0A088AR87 A0A0N0U4H2 E2BSP9 E2AWT9 A0A0C9Q4M4 F4W9K1 A0A158NJF5 A0A151WI43 A0A195DL19 A0A3L8DUS3 A0A232EEZ4 A0A2J7Q3N5 A0A0L0BQI8 A0A0A1WZ30 W8C1E8 A0A1I8P6P0 A0A0K8UEZ4 A0A1I8P6Q1 A0A034VR38 A0A0K8UGJ7 B3M2J8 A0A1I8ND12 A0A0P8Y0H1 B4NLA0 B4KBZ5 A0A182HTJ4 A0A195BHU1 A0A182XIS2 A0A182UNJ9 A0A195F3Y5 A0A0Q9X7Y1 A0A182LBX1 A0A182U6V7 B4JFL3 A0A067QNI2 A0A0M4EPJ2 A0A084W353 A0A0Q9X2R2 A0A1A9ZAC8 A0A1A9VD35 Q9VEV4-2 B4LX42 A0A0R1E2Z8 A0A1W4UIX9 A0A1B0G4T3 B4GFL5 Q9VEV4 A0A1W4UKR1 B4PQZ6 B3P123 A0A1B0AZE5 B4IBV9 Q295W4 A0A0R1E4F7 A0A0Q9WQ14 A0A1W4U778 A0A1W4U7U0 A0A0R1E3N5 A0A3B0KL53 I5APE5 B4QX33 T1J9I9 R7V952 A0A1S3HH86 A0A0L8I5W1 A0A0L8I6S0 A0A2T7PZ30 L7MC62 V5I041 A0A0P6J253 G5BWF2 A0A2Y9LLK4 A0A2Y9LEC0 A0A2Y9HQA9 A0A3Q7MVS5 A0A2Y9LGR7 A0A2U3W0C8 L9KXV9 A0A384AM87 A0A286Y0A2 A0A341BAZ0 A0A3Q7MTS3 A0A286XCM5 A0A3G9GRT0 F6TYT8 A0A3Q7P6M2 A0A2Y9EFK1 A0A2Y9EG90 F1MQK7 A0A384AM70 A0A3Q2HSZ4
Pubmed
EMBL
RSAL01000031
RVE51629.1
ODYU01002073
SOQ39121.1
AGBW02009086
OWR51707.1
+ More
KQ461137 KPJ08821.1 KQ459603 KPI93069.1 KZ288313 PBC28338.1 KQ435837 KOX71482.1 GL450241 EFN81295.1 GL443425 EFN62059.1 GBYB01009038 JAG78805.1 GL888032 EGI69119.1 ADTU01017675 ADTU01017676 KQ983097 KYQ47513.1 KQ980762 KYN13537.1 QOIP01000004 RLU23923.1 NNAY01005163 OXU16930.1 NEVH01019065 PNF23194.1 JRES01001621 KNC21479.1 GBXI01010180 JAD04112.1 GAMC01011101 JAB95454.1 GDHF01027409 JAI24905.1 GAKP01014370 JAC44582.1 GDHF01026527 JAI25787.1 CH902617 EDV43451.1 KPU80275.1 CH964272 EDW84303.1 CH933806 EDW15847.1 APCN01000536 KQ976480 KYM83753.1 KQ981826 KYN35101.1 KRG01754.1 CH916369 EDV93494.1 KK853134 KDR10859.1 CP012526 ALC47715.1 ATLV01019816 ATLV01019817 KE525279 KFB44647.1 KRG01753.1 AE014297 AY121652 BT021249 AAM51979.1 AAX33397.1 CH940650 EDW66694.1 CM000160 KRK03648.1 CCAG010006284 CH479182 EDW34400.1 EDW97318.1 CH954181 EDV49142.1 JXJN01006255 CH480827 EDW44867.1 CM000070 EAL28594.1 KRK03646.1 KRF82861.1 KRF82862.1 KRK03647.1 OUUW01000008 SPP84538.1 EIM52830.2 CM000364 EDX12687.1 JH431975 AMQN01005466 KB296106 ELU12250.1 KQ416499 KOF96729.1 KOF96730.1 PZQS01000001 PVD38671.1 GACK01003602 JAA61432.1 GANP01002156 JAB82312.1 GEBF01006308 JAN97324.1 JH172216 EHB13613.1 KB320604 ELW67636.1 AAKN02007516 LC192295 BBG62335.1
KQ461137 KPJ08821.1 KQ459603 KPI93069.1 KZ288313 PBC28338.1 KQ435837 KOX71482.1 GL450241 EFN81295.1 GL443425 EFN62059.1 GBYB01009038 JAG78805.1 GL888032 EGI69119.1 ADTU01017675 ADTU01017676 KQ983097 KYQ47513.1 KQ980762 KYN13537.1 QOIP01000004 RLU23923.1 NNAY01005163 OXU16930.1 NEVH01019065 PNF23194.1 JRES01001621 KNC21479.1 GBXI01010180 JAD04112.1 GAMC01011101 JAB95454.1 GDHF01027409 JAI24905.1 GAKP01014370 JAC44582.1 GDHF01026527 JAI25787.1 CH902617 EDV43451.1 KPU80275.1 CH964272 EDW84303.1 CH933806 EDW15847.1 APCN01000536 KQ976480 KYM83753.1 KQ981826 KYN35101.1 KRG01754.1 CH916369 EDV93494.1 KK853134 KDR10859.1 CP012526 ALC47715.1 ATLV01019816 ATLV01019817 KE525279 KFB44647.1 KRG01753.1 AE014297 AY121652 BT021249 AAM51979.1 AAX33397.1 CH940650 EDW66694.1 CM000160 KRK03648.1 CCAG010006284 CH479182 EDW34400.1 EDW97318.1 CH954181 EDV49142.1 JXJN01006255 CH480827 EDW44867.1 CM000070 EAL28594.1 KRK03646.1 KRF82861.1 KRF82862.1 KRK03647.1 OUUW01000008 SPP84538.1 EIM52830.2 CM000364 EDX12687.1 JH431975 AMQN01005466 KB296106 ELU12250.1 KQ416499 KOF96729.1 KOF96730.1 PZQS01000001 PVD38671.1 GACK01003602 JAA61432.1 GANP01002156 JAB82312.1 GEBF01006308 JAN97324.1 JH172216 EHB13613.1 KB320604 ELW67636.1 AAKN02007516 LC192295 BBG62335.1
Proteomes
UP000283053
UP000007151
UP000053240
UP000053268
UP000242457
UP000005203
+ More
UP000053105 UP000008237 UP000000311 UP000007755 UP000005205 UP000075809 UP000078492 UP000279307 UP000215335 UP000235965 UP000037069 UP000095300 UP000007801 UP000095301 UP000007798 UP000009192 UP000075840 UP000078540 UP000076407 UP000075903 UP000078541 UP000075882 UP000075902 UP000001070 UP000027135 UP000092553 UP000030765 UP000092445 UP000078200 UP000000803 UP000008792 UP000002282 UP000192221 UP000092444 UP000008744 UP000008711 UP000092460 UP000001292 UP000001819 UP000268350 UP000000304 UP000014760 UP000085678 UP000053454 UP000245119 UP000006813 UP000248483 UP000248481 UP000286641 UP000245340 UP000011518 UP000261681 UP000005447 UP000252040 UP000002281 UP000009136
UP000053105 UP000008237 UP000000311 UP000007755 UP000005205 UP000075809 UP000078492 UP000279307 UP000215335 UP000235965 UP000037069 UP000095300 UP000007801 UP000095301 UP000007798 UP000009192 UP000075840 UP000078540 UP000076407 UP000075903 UP000078541 UP000075882 UP000075902 UP000001070 UP000027135 UP000092553 UP000030765 UP000092445 UP000078200 UP000000803 UP000008792 UP000002282 UP000192221 UP000092444 UP000008744 UP000008711 UP000092460 UP000001292 UP000001819 UP000268350 UP000000304 UP000014760 UP000085678 UP000053454 UP000245119 UP000006813 UP000248483 UP000248481 UP000286641 UP000245340 UP000011518 UP000261681 UP000005447 UP000252040 UP000002281 UP000009136
Interpro
SUPFAM
SSF48371
SSF48371
ProteinModelPortal
A0A3S2NMS2
A0A2H1VEA9
A0A212FDA9
A0A194QYZ8
A0A194PK18
A0A2A3E9D9
+ More
A0A088AR87 A0A0N0U4H2 E2BSP9 E2AWT9 A0A0C9Q4M4 F4W9K1 A0A158NJF5 A0A151WI43 A0A195DL19 A0A3L8DUS3 A0A232EEZ4 A0A2J7Q3N5 A0A0L0BQI8 A0A0A1WZ30 W8C1E8 A0A1I8P6P0 A0A0K8UEZ4 A0A1I8P6Q1 A0A034VR38 A0A0K8UGJ7 B3M2J8 A0A1I8ND12 A0A0P8Y0H1 B4NLA0 B4KBZ5 A0A182HTJ4 A0A195BHU1 A0A182XIS2 A0A182UNJ9 A0A195F3Y5 A0A0Q9X7Y1 A0A182LBX1 A0A182U6V7 B4JFL3 A0A067QNI2 A0A0M4EPJ2 A0A084W353 A0A0Q9X2R2 A0A1A9ZAC8 A0A1A9VD35 Q9VEV4-2 B4LX42 A0A0R1E2Z8 A0A1W4UIX9 A0A1B0G4T3 B4GFL5 Q9VEV4 A0A1W4UKR1 B4PQZ6 B3P123 A0A1B0AZE5 B4IBV9 Q295W4 A0A0R1E4F7 A0A0Q9WQ14 A0A1W4U778 A0A1W4U7U0 A0A0R1E3N5 A0A3B0KL53 I5APE5 B4QX33 T1J9I9 R7V952 A0A1S3HH86 A0A0L8I5W1 A0A0L8I6S0 A0A2T7PZ30 L7MC62 V5I041 A0A0P6J253 G5BWF2 A0A2Y9LLK4 A0A2Y9LEC0 A0A2Y9HQA9 A0A3Q7MVS5 A0A2Y9LGR7 A0A2U3W0C8 L9KXV9 A0A384AM87 A0A286Y0A2 A0A341BAZ0 A0A3Q7MTS3 A0A286XCM5 A0A3G9GRT0 F6TYT8 A0A3Q7P6M2 A0A2Y9EFK1 A0A2Y9EG90 F1MQK7 A0A384AM70 A0A3Q2HSZ4
A0A088AR87 A0A0N0U4H2 E2BSP9 E2AWT9 A0A0C9Q4M4 F4W9K1 A0A158NJF5 A0A151WI43 A0A195DL19 A0A3L8DUS3 A0A232EEZ4 A0A2J7Q3N5 A0A0L0BQI8 A0A0A1WZ30 W8C1E8 A0A1I8P6P0 A0A0K8UEZ4 A0A1I8P6Q1 A0A034VR38 A0A0K8UGJ7 B3M2J8 A0A1I8ND12 A0A0P8Y0H1 B4NLA0 B4KBZ5 A0A182HTJ4 A0A195BHU1 A0A182XIS2 A0A182UNJ9 A0A195F3Y5 A0A0Q9X7Y1 A0A182LBX1 A0A182U6V7 B4JFL3 A0A067QNI2 A0A0M4EPJ2 A0A084W353 A0A0Q9X2R2 A0A1A9ZAC8 A0A1A9VD35 Q9VEV4-2 B4LX42 A0A0R1E2Z8 A0A1W4UIX9 A0A1B0G4T3 B4GFL5 Q9VEV4 A0A1W4UKR1 B4PQZ6 B3P123 A0A1B0AZE5 B4IBV9 Q295W4 A0A0R1E4F7 A0A0Q9WQ14 A0A1W4U778 A0A1W4U7U0 A0A0R1E3N5 A0A3B0KL53 I5APE5 B4QX33 T1J9I9 R7V952 A0A1S3HH86 A0A0L8I5W1 A0A0L8I6S0 A0A2T7PZ30 L7MC62 V5I041 A0A0P6J253 G5BWF2 A0A2Y9LLK4 A0A2Y9LEC0 A0A2Y9HQA9 A0A3Q7MVS5 A0A2Y9LGR7 A0A2U3W0C8 L9KXV9 A0A384AM87 A0A286Y0A2 A0A341BAZ0 A0A3Q7MTS3 A0A286XCM5 A0A3G9GRT0 F6TYT8 A0A3Q7P6M2 A0A2Y9EFK1 A0A2Y9EG90 F1MQK7 A0A384AM70 A0A3Q2HSZ4
Ontologies
GO
PANTHER
Topology
Subcellular location
Cytoplasmic vesicle
Under normal conditions detected in small punctate structures throughout the cytosol of fat body cells, upon induction of autophagy by starvation becomes localized to the outer membrane surface of autophagosomes. Not detected in the Golgi of Garland cells. With evidence from 4 publications.
Autophagosome membrane Under normal conditions detected in small punctate structures throughout the cytosol of fat body cells, upon induction of autophagy by starvation becomes localized to the outer membrane surface of autophagosomes. Not detected in the Golgi of Garland cells. With evidence from 4 publications.
Late endosome membrane Under normal conditions detected in small punctate structures throughout the cytosol of fat body cells, upon induction of autophagy by starvation becomes localized to the outer membrane surface of autophagosomes. Not detected in the Golgi of Garland cells. With evidence from 4 publications.
Golgi apparatus membrane Under normal conditions detected in small punctate structures throughout the cytosol of fat body cells, upon induction of autophagy by starvation becomes localized to the outer membrane surface of autophagosomes. Not detected in the Golgi of Garland cells. With evidence from 4 publications.
Autophagosome membrane Under normal conditions detected in small punctate structures throughout the cytosol of fat body cells, upon induction of autophagy by starvation becomes localized to the outer membrane surface of autophagosomes. Not detected in the Golgi of Garland cells. With evidence from 4 publications.
Late endosome membrane Under normal conditions detected in small punctate structures throughout the cytosol of fat body cells, upon induction of autophagy by starvation becomes localized to the outer membrane surface of autophagosomes. Not detected in the Golgi of Garland cells. With evidence from 4 publications.
Golgi apparatus membrane Under normal conditions detected in small punctate structures throughout the cytosol of fat body cells, upon induction of autophagy by starvation becomes localized to the outer membrane surface of autophagosomes. Not detected in the Golgi of Garland cells. With evidence from 4 publications.
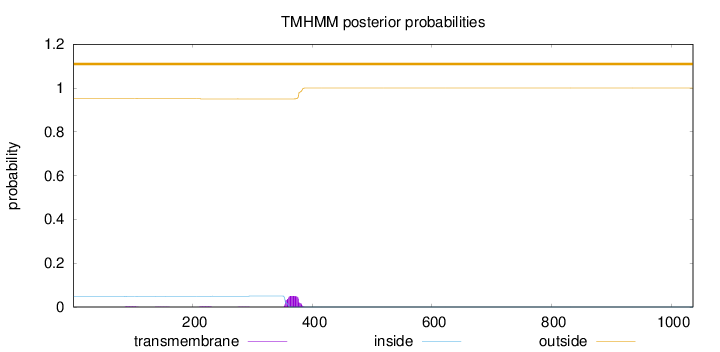
Length:
1036
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.21811
Exp number, first 60 AAs:
0
Total prob of N-in:
0.04863
outside
1 - 1036
Population Genetic Test Statistics
Pi
244.355585
Theta
179.418928
Tajima's D
0.299646
CLR
0.451735
CSRT
0.457227138643068
Interpretation
Uncertain
