Gene
KWMTBOMO10455
Pre Gene Modal
BGIBMGA007005
Annotation
PREDICTED:_apyrase_[Papilio_polytes]
Location in the cell
Cytoplasmic Reliability : 2.505
Sequence
CDS
ATGTATTCACATAAAGGGCGTGGTATGGAATTGTCTGAACTAATTGTTTATGATGGTAGACTGCTCACATTTGATGATCGATCAGGCATGGTTTTTGAAATAATATCTAACAAAATGGTGCCATGGTTAGTGTTGACTGATGGTAATGGTCATGTCGAAAAAGGCTTTAAATCAGAGTGGGCCGCAATGAAAGATGAAATTCTATACATAGGATCTATGGGCAAAGAATGGACAACTTCCAGTGGAGAGTTTGAGAACTATGACCCAATGTGGGTAAAAGCTGTTAATATAAATGGAGAGGTTCAACATTTGACATGGGTAAATCGATATAAAGCCATTAGAAGTTCAATCGGGGTACAATGGCCTGGTTATGTTATACACGAGTCAGGAGTCTGGTCACCTCTCAAGCGACTATGGCATTTCCTGCCGAGGAGGTGTAGTTACGAACAATACAATGAAACAAAGGACGAAATTAAGGGTTGCAACTACCTTATTACTGCAGATGATAATTTCAGAAATATTAAAGCTAACAAGATTACAAAATTTCAACCTAAGCATGGCTTCTCATCGTTCAAGTTTATACCTGGATCTAATGATGAGGTCATAGTGGCTTTGAAAACAACTGAGTTTGAAGGCAAGACCGCAACATACATAACAGCATTCACCACTGATGGCTTAGAGTTACTATCCGATACTTTTGTGGAAAACATGAAATATGAGGGCATCGAGTTTTTGTGA
Protein
MYSHKGRGMELSELIVYDGRLLTFDDRSGMVFEIISNKMVPWLVLTDGNGHVEKGFKSEWAAMKDEILYIGSMGKEWTTSSGEFENYDPMWVKAVNINGEVQHLTWVNRYKAIRSSIGVQWPGYVIHESGVWSPLKRLWHFLPRRCSYEQYNETKDEIKGCNYLITADDNFRNIKANKITKFQPKHGFSSFKFIPGSNDEVIVALKTTEFEGKTATYITAFTTDGLELLSDTFVENMKYEGIEFL
Summary
Uniprot
H9JBW0
A0A2H1VD75
A0A194QU28
A0A194PIC7
A0A2A4J103
I4DPR5
+ More
A0A3S2P3S4 A0A212FE02 A0A1E1W0B6 S4PS75 D6WUT9 A0A182K7E3 A0A084W023 A0A182PRV7 W5J8C2 A0A182UXL3 Q7PX52 A0A182HKB3 A0A182TVI5 A0A182M482 A0A182L8J6 A0A2M4BQ49 A0A182X9C6 A0A2M4BNI2 A0A2M4BPC5 A0A2M4A8X0 A0A2M4A906 U5EG83 A0A182QAS1 A0A182FS33 A0A182YHX0 A0A182RSD7 A0A2M3Z432 A0A0K8TT62 A0A2M3Z3A2 A0A2M3Z3K5 A0A182NQS5 A0A023ES30 U4UKX4 N6U339 A0A182HAM8 Q175I8 A0A1W4X9M1 A0A336MT34 A0A232FNA1 A0A3L8DRR4 A0A026VZ92 E2B590 B4HI25 K7IWR2 A0A151WZ85 A0A0T6AU84 A0A195F525 B3LXB8 A0A195D4X7 F4WXT2 A0A1B0CNJ1 A0A1W4WD12 A0A195BUD7 A0A1L8DXI2 A0A195EJA6 A0A158NT68 A0A0M3QXE8 A0A1L8DXM0 E9IHR8 A0AN69 A0AN64 B4NAX1 A0A1Q3F8H0 A0AN61 Q9VGN8 A0AN66 B3NZE8 A0A1Q3F8C8 A0A2A3E1Z0 E2B1L5 A0A088ATM5 A0A0J7KNW6 A0A1Y1LB92 B4QU37 A0AN67 A0A1Y1L987 Q295A5 B4GMD4 B4PM57 A0A182VW10 R4WDI7 A0A3B0KWJ7 B4M5C7 A0A0K8UV76 B4JUN8 A0A1B0CG27 A0A0A9YZ34 A0A034V8W1 A0A0K8SGY6 W8APQ5 A0A0A1WZY5 A3EXU1 B4KDY9 A0A1B6MBE8
A0A3S2P3S4 A0A212FE02 A0A1E1W0B6 S4PS75 D6WUT9 A0A182K7E3 A0A084W023 A0A182PRV7 W5J8C2 A0A182UXL3 Q7PX52 A0A182HKB3 A0A182TVI5 A0A182M482 A0A182L8J6 A0A2M4BQ49 A0A182X9C6 A0A2M4BNI2 A0A2M4BPC5 A0A2M4A8X0 A0A2M4A906 U5EG83 A0A182QAS1 A0A182FS33 A0A182YHX0 A0A182RSD7 A0A2M3Z432 A0A0K8TT62 A0A2M3Z3A2 A0A2M3Z3K5 A0A182NQS5 A0A023ES30 U4UKX4 N6U339 A0A182HAM8 Q175I8 A0A1W4X9M1 A0A336MT34 A0A232FNA1 A0A3L8DRR4 A0A026VZ92 E2B590 B4HI25 K7IWR2 A0A151WZ85 A0A0T6AU84 A0A195F525 B3LXB8 A0A195D4X7 F4WXT2 A0A1B0CNJ1 A0A1W4WD12 A0A195BUD7 A0A1L8DXI2 A0A195EJA6 A0A158NT68 A0A0M3QXE8 A0A1L8DXM0 E9IHR8 A0AN69 A0AN64 B4NAX1 A0A1Q3F8H0 A0AN61 Q9VGN8 A0AN66 B3NZE8 A0A1Q3F8C8 A0A2A3E1Z0 E2B1L5 A0A088ATM5 A0A0J7KNW6 A0A1Y1LB92 B4QU37 A0AN67 A0A1Y1L987 Q295A5 B4GMD4 B4PM57 A0A182VW10 R4WDI7 A0A3B0KWJ7 B4M5C7 A0A0K8UV76 B4JUN8 A0A1B0CG27 A0A0A9YZ34 A0A034V8W1 A0A0K8SGY6 W8APQ5 A0A0A1WZY5 A3EXU1 B4KDY9 A0A1B6MBE8
Pubmed
19121390
26354079
22651552
22118469
23622113
18362917
+ More
19820115 24438588 20920257 23761445 12364791 14747013 17210077 20966253 25244985 26369729 24945155 23537049 26483478 17510324 28648823 30249741 24508170 20798317 17994087 20075255 21719571 21347285 21282665 16951084 19126864 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28004739 15632085 17550304 23691247 25401762 25348373 26823975 24495485 25830018
19820115 24438588 20920257 23761445 12364791 14747013 17210077 20966253 25244985 26369729 24945155 23537049 26483478 17510324 28648823 30249741 24508170 20798317 17994087 20075255 21719571 21347285 21282665 16951084 19126864 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28004739 15632085 17550304 23691247 25401762 25348373 26823975 24495485 25830018
EMBL
BABH01014900
ODYU01001657
SOQ38184.1
KQ461137
KPJ08819.1
KQ459603
+ More
KPI93067.1 NWSH01003936 PCG65661.1 AK403738 BAM19905.1 RSAL01000031 RVE51631.1 AGBW02008995 OWR51938.1 GDQN01010638 GDQN01005784 JAT80416.1 JAT85270.1 GAIX01000185 JAA92375.1 KQ971363 EFA07795.2 ATLV01019081 KE525262 KFB43567.1 ADMH02002101 ETN59125.1 AAAB01008987 EAA01797.3 APCN01000824 AXCM01006044 GGFJ01005993 MBW55134.1 GGFJ01005496 MBW54637.1 GGFJ01005497 MBW54638.1 GGFK01003880 MBW37201.1 GGFK01003889 MBW37210.1 GANO01003550 JAB56321.1 AXCN02001500 GGFM01002523 MBW23274.1 GDAI01000260 JAI17343.1 GGFM01002241 MBW22992.1 GGFM01002346 MBW23097.1 GAPW01001558 JAC12040.1 KB632255 ERL90655.1 APGK01041593 APGK01041594 KB740994 ENN75985.1 JXUM01122999 KQ566666 KXJ69928.1 CH477400 EAT41732.1 UFQS01002181 UFQT01002181 SSX13434.1 SSX32865.1 NNAY01000010 OXU32053.1 QOIP01000005 RLU22488.1 KK107570 EZA48791.1 GL445739 EFN89143.1 CH480815 EDW42607.1 AAZX01000616 KQ982649 KYQ53203.1 LJIG01022799 KRT78652.1 KQ981805 KYN35496.1 CH902617 EDV42762.1 KQ976842 KYN07938.1 GL888427 EGI61010.1 AJWK01020407 KQ976403 KYM92187.1 GFDF01002926 JAV11158.1 KQ978801 KYN28241.1 ADTU01025665 CP012526 ALC45803.1 GFDF01002925 JAV11159.1 GL763294 EFZ19843.1 AM294107 CAL26011.1 AM294102 FM245221 CAL26006.1 CAR93147.1 CH964232 EDW80935.1 GFDL01011257 JAV23788.1 AM294099 AM294100 AM294101 AM294103 AM294108 FM245215 FM245216 FM245219 CAL26003.1 CAL26004.1 CAL26005.1 CAL26007.1 CAL26012.1 CAR93141.1 CAR93142.1 CAR93145.1 AE014297 AY058610 AM294097 AM294098 AM294106 FM245213 FM245214 FM245217 FM245218 FM245220 FM245222 FM245223 FM245224 AAF54638.1 AAL13839.1 CAL26001.1 CAL26002.1 CAL26010.1 CAR93139.1 CAR93140.1 CAR93143.1 CAR93144.1 CAR93146.1 CAR93148.1 CAR93149.1 CAR93150.1 AM294104 CAL26008.1 CH954181 EDV49521.1 GFDL01011258 JAV23787.1 KZ288452 PBC25544.1 GL444953 EFN60371.1 LBMM01004987 KMQ91934.1 GEZM01061964 GEZM01061963 GEZM01061958 JAV70158.1 CM000364 EDX13368.1 AM294105 CAL26009.1 GEZM01061962 GEZM01061961 GEZM01061960 JAV70164.1 CM000070 EAL28807.2 CH479185 EDW38008.1 CM000160 EDW96923.1 AK417738 BAN20953.1 OUUW01000014 SPP88398.1 CH940652 EDW59838.1 GDHF01021823 GDHF01003531 JAI30491.1 JAI48783.1 CH916374 EDV91208.1 AJWK01010665 AJWK01010666 GBHO01016110 GBHO01005337 GBHO01005328 JAG27494.1 JAG38267.1 JAG38276.1 GAKP01020957 GAKP01020956 GAKP01020954 JAC37998.1 GBRD01013270 GBRD01013268 GBRD01010634 GDHC01007054 JAG52558.1 JAQ11575.1 GAMC01015787 GAMC01015785 GAMC01015783 JAB90770.1 GBXI01010202 JAD04090.1 EF092009 ABN12001.1 CH933806 EDW16010.1 GEBQ01006726 JAT33251.1
KPI93067.1 NWSH01003936 PCG65661.1 AK403738 BAM19905.1 RSAL01000031 RVE51631.1 AGBW02008995 OWR51938.1 GDQN01010638 GDQN01005784 JAT80416.1 JAT85270.1 GAIX01000185 JAA92375.1 KQ971363 EFA07795.2 ATLV01019081 KE525262 KFB43567.1 ADMH02002101 ETN59125.1 AAAB01008987 EAA01797.3 APCN01000824 AXCM01006044 GGFJ01005993 MBW55134.1 GGFJ01005496 MBW54637.1 GGFJ01005497 MBW54638.1 GGFK01003880 MBW37201.1 GGFK01003889 MBW37210.1 GANO01003550 JAB56321.1 AXCN02001500 GGFM01002523 MBW23274.1 GDAI01000260 JAI17343.1 GGFM01002241 MBW22992.1 GGFM01002346 MBW23097.1 GAPW01001558 JAC12040.1 KB632255 ERL90655.1 APGK01041593 APGK01041594 KB740994 ENN75985.1 JXUM01122999 KQ566666 KXJ69928.1 CH477400 EAT41732.1 UFQS01002181 UFQT01002181 SSX13434.1 SSX32865.1 NNAY01000010 OXU32053.1 QOIP01000005 RLU22488.1 KK107570 EZA48791.1 GL445739 EFN89143.1 CH480815 EDW42607.1 AAZX01000616 KQ982649 KYQ53203.1 LJIG01022799 KRT78652.1 KQ981805 KYN35496.1 CH902617 EDV42762.1 KQ976842 KYN07938.1 GL888427 EGI61010.1 AJWK01020407 KQ976403 KYM92187.1 GFDF01002926 JAV11158.1 KQ978801 KYN28241.1 ADTU01025665 CP012526 ALC45803.1 GFDF01002925 JAV11159.1 GL763294 EFZ19843.1 AM294107 CAL26011.1 AM294102 FM245221 CAL26006.1 CAR93147.1 CH964232 EDW80935.1 GFDL01011257 JAV23788.1 AM294099 AM294100 AM294101 AM294103 AM294108 FM245215 FM245216 FM245219 CAL26003.1 CAL26004.1 CAL26005.1 CAL26007.1 CAL26012.1 CAR93141.1 CAR93142.1 CAR93145.1 AE014297 AY058610 AM294097 AM294098 AM294106 FM245213 FM245214 FM245217 FM245218 FM245220 FM245222 FM245223 FM245224 AAF54638.1 AAL13839.1 CAL26001.1 CAL26002.1 CAL26010.1 CAR93139.1 CAR93140.1 CAR93143.1 CAR93144.1 CAR93146.1 CAR93148.1 CAR93149.1 CAR93150.1 AM294104 CAL26008.1 CH954181 EDV49521.1 GFDL01011258 JAV23787.1 KZ288452 PBC25544.1 GL444953 EFN60371.1 LBMM01004987 KMQ91934.1 GEZM01061964 GEZM01061963 GEZM01061958 JAV70158.1 CM000364 EDX13368.1 AM294105 CAL26009.1 GEZM01061962 GEZM01061961 GEZM01061960 JAV70164.1 CM000070 EAL28807.2 CH479185 EDW38008.1 CM000160 EDW96923.1 AK417738 BAN20953.1 OUUW01000014 SPP88398.1 CH940652 EDW59838.1 GDHF01021823 GDHF01003531 JAI30491.1 JAI48783.1 CH916374 EDV91208.1 AJWK01010665 AJWK01010666 GBHO01016110 GBHO01005337 GBHO01005328 JAG27494.1 JAG38267.1 JAG38276.1 GAKP01020957 GAKP01020956 GAKP01020954 JAC37998.1 GBRD01013270 GBRD01013268 GBRD01010634 GDHC01007054 JAG52558.1 JAQ11575.1 GAMC01015787 GAMC01015785 GAMC01015783 JAB90770.1 GBXI01010202 JAD04090.1 EF092009 ABN12001.1 CH933806 EDW16010.1 GEBQ01006726 JAT33251.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000283053
UP000007151
+ More
UP000007266 UP000075881 UP000030765 UP000075885 UP000000673 UP000075903 UP000007062 UP000075840 UP000075902 UP000075883 UP000075882 UP000076407 UP000075886 UP000069272 UP000076408 UP000075900 UP000075884 UP000030742 UP000019118 UP000069940 UP000249989 UP000008820 UP000192223 UP000215335 UP000279307 UP000053097 UP000008237 UP000001292 UP000002358 UP000075809 UP000078541 UP000007801 UP000078542 UP000007755 UP000092461 UP000192221 UP000078540 UP000078492 UP000005205 UP000092553 UP000007798 UP000000803 UP000008711 UP000242457 UP000000311 UP000005203 UP000036403 UP000000304 UP000001819 UP000008744 UP000002282 UP000075920 UP000268350 UP000008792 UP000001070 UP000009192
UP000007266 UP000075881 UP000030765 UP000075885 UP000000673 UP000075903 UP000007062 UP000075840 UP000075902 UP000075883 UP000075882 UP000076407 UP000075886 UP000069272 UP000076408 UP000075900 UP000075884 UP000030742 UP000019118 UP000069940 UP000249989 UP000008820 UP000192223 UP000215335 UP000279307 UP000053097 UP000008237 UP000001292 UP000002358 UP000075809 UP000078541 UP000007801 UP000078542 UP000007755 UP000092461 UP000192221 UP000078540 UP000078492 UP000005205 UP000092553 UP000007798 UP000000803 UP000008711 UP000242457 UP000000311 UP000005203 UP000036403 UP000000304 UP000001819 UP000008744 UP000002282 UP000075920 UP000268350 UP000008792 UP000001070 UP000009192
Interpro
IPR036258
Apyrase_sf
+ More
IPR009283 Apyrase
IPR019412 Iml2/TPR_39
IPR011990 TPR-like_helical_dom_sf
IPR000897 SRP54_GTPase_dom
IPR027417 P-loop_NTPase
IPR013822 Signal_recog_particl_SRP54_hlx
IPR003593 AAA+_ATPase
IPR036891 Signal_recog_part_SRP54_M_sf
IPR036225 SRP/SRP_N
IPR004125 Signal_recog_particle_SRP54_M
IPR022941 SRP54
IPR042101 SRP54_N_sf
IPR009283 Apyrase
IPR019412 Iml2/TPR_39
IPR011990 TPR-like_helical_dom_sf
IPR000897 SRP54_GTPase_dom
IPR027417 P-loop_NTPase
IPR013822 Signal_recog_particl_SRP54_hlx
IPR003593 AAA+_ATPase
IPR036891 Signal_recog_part_SRP54_M_sf
IPR036225 SRP/SRP_N
IPR004125 Signal_recog_particle_SRP54_M
IPR022941 SRP54
IPR042101 SRP54_N_sf
SUPFAM
Gene 3D
ProteinModelPortal
H9JBW0
A0A2H1VD75
A0A194QU28
A0A194PIC7
A0A2A4J103
I4DPR5
+ More
A0A3S2P3S4 A0A212FE02 A0A1E1W0B6 S4PS75 D6WUT9 A0A182K7E3 A0A084W023 A0A182PRV7 W5J8C2 A0A182UXL3 Q7PX52 A0A182HKB3 A0A182TVI5 A0A182M482 A0A182L8J6 A0A2M4BQ49 A0A182X9C6 A0A2M4BNI2 A0A2M4BPC5 A0A2M4A8X0 A0A2M4A906 U5EG83 A0A182QAS1 A0A182FS33 A0A182YHX0 A0A182RSD7 A0A2M3Z432 A0A0K8TT62 A0A2M3Z3A2 A0A2M3Z3K5 A0A182NQS5 A0A023ES30 U4UKX4 N6U339 A0A182HAM8 Q175I8 A0A1W4X9M1 A0A336MT34 A0A232FNA1 A0A3L8DRR4 A0A026VZ92 E2B590 B4HI25 K7IWR2 A0A151WZ85 A0A0T6AU84 A0A195F525 B3LXB8 A0A195D4X7 F4WXT2 A0A1B0CNJ1 A0A1W4WD12 A0A195BUD7 A0A1L8DXI2 A0A195EJA6 A0A158NT68 A0A0M3QXE8 A0A1L8DXM0 E9IHR8 A0AN69 A0AN64 B4NAX1 A0A1Q3F8H0 A0AN61 Q9VGN8 A0AN66 B3NZE8 A0A1Q3F8C8 A0A2A3E1Z0 E2B1L5 A0A088ATM5 A0A0J7KNW6 A0A1Y1LB92 B4QU37 A0AN67 A0A1Y1L987 Q295A5 B4GMD4 B4PM57 A0A182VW10 R4WDI7 A0A3B0KWJ7 B4M5C7 A0A0K8UV76 B4JUN8 A0A1B0CG27 A0A0A9YZ34 A0A034V8W1 A0A0K8SGY6 W8APQ5 A0A0A1WZY5 A3EXU1 B4KDY9 A0A1B6MBE8
A0A3S2P3S4 A0A212FE02 A0A1E1W0B6 S4PS75 D6WUT9 A0A182K7E3 A0A084W023 A0A182PRV7 W5J8C2 A0A182UXL3 Q7PX52 A0A182HKB3 A0A182TVI5 A0A182M482 A0A182L8J6 A0A2M4BQ49 A0A182X9C6 A0A2M4BNI2 A0A2M4BPC5 A0A2M4A8X0 A0A2M4A906 U5EG83 A0A182QAS1 A0A182FS33 A0A182YHX0 A0A182RSD7 A0A2M3Z432 A0A0K8TT62 A0A2M3Z3A2 A0A2M3Z3K5 A0A182NQS5 A0A023ES30 U4UKX4 N6U339 A0A182HAM8 Q175I8 A0A1W4X9M1 A0A336MT34 A0A232FNA1 A0A3L8DRR4 A0A026VZ92 E2B590 B4HI25 K7IWR2 A0A151WZ85 A0A0T6AU84 A0A195F525 B3LXB8 A0A195D4X7 F4WXT2 A0A1B0CNJ1 A0A1W4WD12 A0A195BUD7 A0A1L8DXI2 A0A195EJA6 A0A158NT68 A0A0M3QXE8 A0A1L8DXM0 E9IHR8 A0AN69 A0AN64 B4NAX1 A0A1Q3F8H0 A0AN61 Q9VGN8 A0AN66 B3NZE8 A0A1Q3F8C8 A0A2A3E1Z0 E2B1L5 A0A088ATM5 A0A0J7KNW6 A0A1Y1LB92 B4QU37 A0AN67 A0A1Y1L987 Q295A5 B4GMD4 B4PM57 A0A182VW10 R4WDI7 A0A3B0KWJ7 B4M5C7 A0A0K8UV76 B4JUN8 A0A1B0CG27 A0A0A9YZ34 A0A034V8W1 A0A0K8SGY6 W8APQ5 A0A0A1WZY5 A3EXU1 B4KDY9 A0A1B6MBE8
PDB
2H2U
E-value=1.21746e-75,
Score=718
Ontologies
PATHWAY
GO
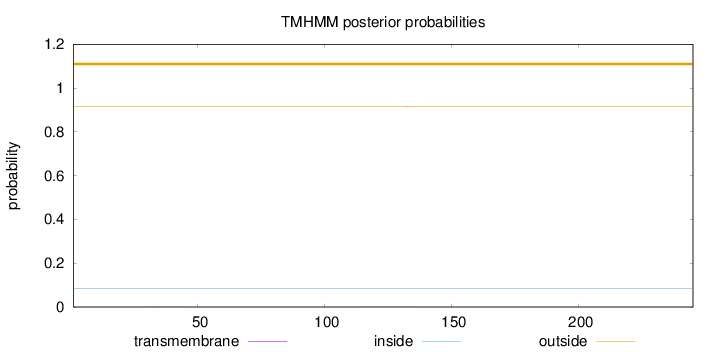
Topology
Length:
245
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01307
Exp number, first 60 AAs:
0.00454
Total prob of N-in:
0.08365
outside
1 - 245
Population Genetic Test Statistics
Pi
218.535491
Theta
149.342623
Tajima's D
-0.88103
CLR
1.324367
CSRT
0.160341982900855
Interpretation
Uncertain
