Gene
KWMTBOMO10440
Pre Gene Modal
BGIBMGA007089
Annotation
PREDICTED:_zinc_finger_protein_569-like_isoform_X3_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 3.16
Sequence
CDS
ATGAACCCAGAACACCACAATATTACTAGTGGTGGTCAGCCATCGGGAAGTACAGAGTCTCAAAATCAACGGACAGCTCAGCAACAGCAGCAGACTAATTTAACACCCACAACCTCTGCTACTGACCTCCGAGTGAACTCTGCTGCTGTAAATGTCGCTTTGTCTAGTGTTGCGAAGTACTGGGTGTTTACAAATCTCTTCCCTGGTCCAATACCACAAGTATCAGTGTATGGACTACCAACAGGTACAAGAATAGAAAACGGGAAAGCAGTACAGGATATTGGTCAAACACATACTGGTATACTTAATGGTGATCCTAATATCATACTTGGTCATCATGGAGGTCAGGCACAGGTTACAGTGTCTGCGTCTGGAACACAGCAGATTCCAGTTTCACAAATCCTTTCAACACAATCTGGGCAAACTCATGAAACGATGGTGGGCCACGCTCAGGCACAAGAGCTTGGAGGACAACAGGCGACAGGTAGTGCCGGAGCTGGTCAGGCCACACACCAGCAGGTAGCCAATAGTCGGGTCGAGTTTGTACAACACCATAATATCGATATGACTCACCACACACAACAGCATTTGTTGCAACAACAGTTAATGGCGGCTCGACCAGACCATTCTAATCAACAGATACAACTAACTGTCAGTGAAGATGGTATAGTGACAGTGGTAGAGCCGGGAGCAGGTAAAATTGTTGAAAAGGAAGAGCTTCATGAAGCTATCAAACTACCCAATGATCAAACACTCACTGTACATCAACTACAGCAAATCGTAGGACAACAGGTAATAGACAGCGTGGTTCGCATAGAACAAGCCACGGGAGAACCAGCCAATATTTTAGTGACTCACAATGCAGATGGCACCACGTCCATAGAAGCAAGTGCTGCTGAATCGCTCATCGTGAAAGACGAAAAAAATTCAAAGATTGAAACTGCTCAGTTTGCCATACCTACAGACATTAAAGATATCAAAGGCATTGATTTGAAGGAAAATTTACAGACTGTTGGGGCAATGGGAATGGAAGGGGCCGTCGTCAAGATATCAGCTGGAGCTACCGATCATGACATGCATGCTATGTATAAAGTTAACGTGGAAGATCTCTCACAACTACTAGCATATCATGAAGTCTTTGGGAAATTGAATTCTGATGGACAACCACAACAACAACCCAAGGTTATAACGGAAATAGAAGTTGAGGTAGAAGCAGGTACGAGTGCAGCGATAACTGAACAAGATTCGTCACCTGGACACCATTCTTGTGATATTTGTGGGAAAATATTCCAGTTTAGATATCAACTCATTGTACACAGACGATACCATGGCGAAAGCAAACCTTTTGCATGTTTAGTTTGTGGAGTAGCGTTTGCTAACCCTTTAGAATTATCAAGACACGGAAAATGTCATCTTGCCGGCGACCCCGCGGAACGACACGCCAAAAGAATGACGGCGGACAAGCCATATGTTTGCACAACTTGCCATAAAACCTTTTCGCGAAAGGAACATCTGGATAACCACGTACGAAGTCATACTGGAGAAACACCTTACCGATGTGAATTCTGTGCAAAGACATTCACTAGAAAAGAACACATGGTTAATCACGTCAGGAAACACACCGGCGAAACGCCGCATCGCTGCGACATCTGCAAGAAAAGTTTCACGAGGAAAGAACATTTCATGAACCATGTCATGTGGCACACAGGTGAAACTCCACATCGTTGCGACTTTTGTTCGAAAACATTTACTCGTAAGGAGCATCTTCTCAATCATGTAAGACAACATACTGGAGAATCTCCTCATCGGTGTAACTTCTGCTCCAAATCGTTCACGAGAAGAGAACATTTAGTGAACCACGTGAGGCAACATACCGGCGAAACACCATTCCAGTGTGGATACTGTCCTAAAGCATTCACAAGAAAGGATCATCTCGTAAATCACGTTCGACAACACACAGGGGAATCGCCACACAAGTGCTCTTTCTGCACCAAGTCGTTCACAAGAAAAGAACATCTAACGAACCACGTGCGTCAACACACAGGCGAGTCCCCGCATCGATGTACTTACTGCGCCAAGTCTTTTACAAGGAAAGAACATCTTACTAATCATATCAGACAGCACACCGGCGAAACTCCACACAAGTGTACCTACTGTCCGCGTGCCTTCGCGCGTAAGGAACATCTGAACAATCACATTCGACAACACACCGGCGTCACTCCGCACGCCTGTACTTACTGCAGCAAGACATTCACAAGAAAGGAGCACCTCGTTAACCATGTTCGGCAACACACTGGCGAGACACCATTTAAATGTGGATTTTGCACGAAATCGTTCTCTAGAAAAGAACATTTAACAAATCACATCCATCTACACACTGGAGAAACGCCACACAAATGTCCTTTCTGTACAAAGACGTTTTCGAGAAAAGAACACTTGACCAATCATGTCAGAATACATACTGGTGAATCGCCACATCGTTGTGAATTTTGTCAGAAAACATTTACTCGCAAAGAGCATCTCACCAACCATCTAAAACAGCACACTGGAGACACAGCGCATGCATGTAAAGTTTGTTCCAAGCCTTTTACGAGAAAGGAACACCTTGTCACACACATGAGGTAA
Protein
MNPEHHNITSGGQPSGSTESQNQRTAQQQQQTNLTPTTSATDLRVNSAAVNVALSSVAKYWVFTNLFPGPIPQVSVYGLPTGTRIENGKAVQDIGQTHTGILNGDPNIILGHHGGQAQVTVSASGTQQIPVSQILSTQSGQTHETMVGHAQAQELGGQQATGSAGAGQATHQQVANSRVEFVQHHNIDMTHHTQQHLLQQQLMAARPDHSNQQIQLTVSEDGIVTVVEPGAGKIVEKEELHEAIKLPNDQTLTVHQLQQIVGQQVIDSVVRIEQATGEPANILVTHNADGTTSIEASAAESLIVKDEKNSKIETAQFAIPTDIKDIKGIDLKENLQTVGAMGMEGAVVKISAGATDHDMHAMYKVNVEDLSQLLAYHEVFGKLNSDGQPQQQPKVITEIEVEVEAGTSAAITEQDSSPGHHSCDICGKIFQFRYQLIVHRRYHGESKPFACLVCGVAFANPLELSRHGKCHLAGDPAERHAKRMTADKPYVCTTCHKTFSRKEHLDNHVRSHTGETPYRCEFCAKTFTRKEHMVNHVRKHTGETPHRCDICKKSFTRKEHFMNHVMWHTGETPHRCDFCSKTFTRKEHLLNHVRQHTGESPHRCNFCSKSFTRREHLVNHVRQHTGETPFQCGYCPKAFTRKDHLVNHVRQHTGESPHKCSFCTKSFTRKEHLTNHVRQHTGESPHRCTYCAKSFTRKEHLTNHIRQHTGETPHKCTYCPRAFARKEHLNNHIRQHTGVTPHACTYCSKTFTRKEHLVNHVRQHTGETPFKCGFCTKSFSRKEHLTNHIHLHTGETPHKCPFCTKTFSRKEHLTNHVRIHTGESPHRCEFCQKTFTRKEHLTNHLKQHTGDTAHACKVCSKPFTRKEHLVTHMR
Summary
Uniprot
H9JC44
A0A2A4IYQ4
A0A1E1WS31
A0A2A4IZV4
A0A1E1W290
A0A2A4IZN7
+ More
A0A1E1WV67 A0A1E1W8B5 A0A212ERB0 A0A194PIA9 A0A194QU13 A0A2H1X2Y2 A0A1Y1MEC5 D6WYI1 A0A0T6AYN7 A0A1W4XGI0 A0A1W4XS30 E0VFL0 V5I9P9 A0A067RK94 A0A0C9Q4T0 F4W5X7 A0A158P2W0 A0A151XH71 A0A3L8DFL0 V9IF05 A0A154PIG5 A0A0L7R540 V9IEJ4 A0A232F5M4 N6U8B6 A0A0N0BKL8 A0A310SD71 K7IZH9 A0A0C9RPJ2 A0A1B0CTZ0 A0A195CY15 A0A195DFJ8 A0A2P8XG51 A0A088AQW3 A0A0C9RLC9
A0A1E1WV67 A0A1E1W8B5 A0A212ERB0 A0A194PIA9 A0A194QU13 A0A2H1X2Y2 A0A1Y1MEC5 D6WYI1 A0A0T6AYN7 A0A1W4XGI0 A0A1W4XS30 E0VFL0 V5I9P9 A0A067RK94 A0A0C9Q4T0 F4W5X7 A0A158P2W0 A0A151XH71 A0A3L8DFL0 V9IF05 A0A154PIG5 A0A0L7R540 V9IEJ4 A0A232F5M4 N6U8B6 A0A0N0BKL8 A0A310SD71 K7IZH9 A0A0C9RPJ2 A0A1B0CTZ0 A0A195CY15 A0A195DFJ8 A0A2P8XG51 A0A088AQW3 A0A0C9RLC9
Pubmed
EMBL
BABH01014857
BABH01014858
NWSH01004552
PCG64955.1
GDQN01001219
JAT89835.1
+ More
PCG64958.1 GDQN01009971 JAT81083.1 PCG64956.1 GDQN01000172 JAT90882.1 GDQN01007855 JAT83199.1 AGBW02013074 OWR44015.1 KQ459603 KPI93052.1 KQ461137 KPJ08804.1 ODYU01012995 SOQ59586.1 GEZM01033751 GEZM01033750 JAV84179.1 KQ971362 EFA08986.1 LJIG01022489 KRT80303.1 DS235115 EEB12166.1 GALX01002555 JAB65911.1 KK852421 KDR24286.1 GBYB01009088 JAG78855.1 GL887695 EGI70453.1 ADTU01007511 ADTU01007512 KQ982130 KYQ59754.1 QOIP01000009 RLU18953.1 JR040253 AEY59051.1 KQ434924 KZC11602.1 KQ414657 KOC65866.1 JR040254 AEY59052.1 NNAY01000947 OXU25779.1 APGK01035666 KB740928 ENN77910.1 KQ435698 KOX80761.1 KQ762903 OAD55274.1 GBYB01009086 JAG78853.1 AJWK01028174 AJWK01028175 KQ977185 KYN05044.1 KQ980903 KYN11658.1 PYGN01002258 PSN30986.1 GBYB01009085 JAG78852.1
PCG64958.1 GDQN01009971 JAT81083.1 PCG64956.1 GDQN01000172 JAT90882.1 GDQN01007855 JAT83199.1 AGBW02013074 OWR44015.1 KQ459603 KPI93052.1 KQ461137 KPJ08804.1 ODYU01012995 SOQ59586.1 GEZM01033751 GEZM01033750 JAV84179.1 KQ971362 EFA08986.1 LJIG01022489 KRT80303.1 DS235115 EEB12166.1 GALX01002555 JAB65911.1 KK852421 KDR24286.1 GBYB01009088 JAG78855.1 GL887695 EGI70453.1 ADTU01007511 ADTU01007512 KQ982130 KYQ59754.1 QOIP01000009 RLU18953.1 JR040253 AEY59051.1 KQ434924 KZC11602.1 KQ414657 KOC65866.1 JR040254 AEY59052.1 NNAY01000947 OXU25779.1 APGK01035666 KB740928 ENN77910.1 KQ435698 KOX80761.1 KQ762903 OAD55274.1 GBYB01009086 JAG78853.1 AJWK01028174 AJWK01028175 KQ977185 KYN05044.1 KQ980903 KYN11658.1 PYGN01002258 PSN30986.1 GBYB01009085 JAG78852.1
Proteomes
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JC44
A0A2A4IYQ4
A0A1E1WS31
A0A2A4IZV4
A0A1E1W290
A0A2A4IZN7
+ More
A0A1E1WV67 A0A1E1W8B5 A0A212ERB0 A0A194PIA9 A0A194QU13 A0A2H1X2Y2 A0A1Y1MEC5 D6WYI1 A0A0T6AYN7 A0A1W4XGI0 A0A1W4XS30 E0VFL0 V5I9P9 A0A067RK94 A0A0C9Q4T0 F4W5X7 A0A158P2W0 A0A151XH71 A0A3L8DFL0 V9IF05 A0A154PIG5 A0A0L7R540 V9IEJ4 A0A232F5M4 N6U8B6 A0A0N0BKL8 A0A310SD71 K7IZH9 A0A0C9RPJ2 A0A1B0CTZ0 A0A195CY15 A0A195DFJ8 A0A2P8XG51 A0A088AQW3 A0A0C9RLC9
A0A1E1WV67 A0A1E1W8B5 A0A212ERB0 A0A194PIA9 A0A194QU13 A0A2H1X2Y2 A0A1Y1MEC5 D6WYI1 A0A0T6AYN7 A0A1W4XGI0 A0A1W4XS30 E0VFL0 V5I9P9 A0A067RK94 A0A0C9Q4T0 F4W5X7 A0A158P2W0 A0A151XH71 A0A3L8DFL0 V9IF05 A0A154PIG5 A0A0L7R540 V9IEJ4 A0A232F5M4 N6U8B6 A0A0N0BKL8 A0A310SD71 K7IZH9 A0A0C9RPJ2 A0A1B0CTZ0 A0A195CY15 A0A195DFJ8 A0A2P8XG51 A0A088AQW3 A0A0C9RLC9
PDB
5V3M
E-value=5.77101e-61,
Score=597
Ontologies
GO
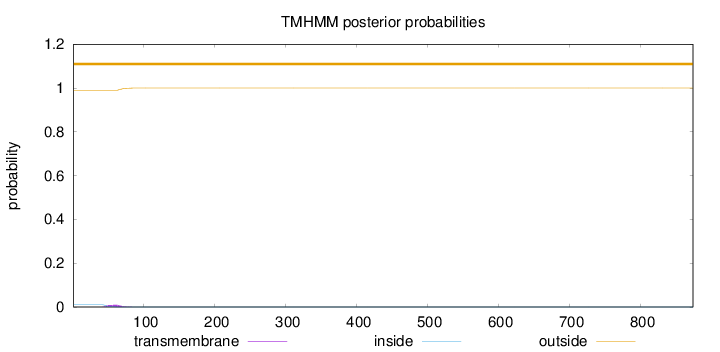
Topology
Length:
874
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.2181
Exp number, first 60 AAs:
0.12989
Total prob of N-in:
0.01037
outside
1 - 874
Population Genetic Test Statistics
Pi
223.419146
Theta
179.226052
Tajima's D
0.219735
CLR
0.90478
CSRT
0.431528423578821
Interpretation
Uncertain
