Pre Gene Modal
BGIBMGA007087
Annotation
uncharacterized_protein_LOC778505_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 3.827
Sequence
CDS
ATGGCCATCAGCAGGTTGTCTATCATCAAGTTTCTGGAGCTGGCACTTACGTGTTCGTGCGTGGCTCTCCACTACCACAGTTACAATGCAGATGCGGATATCGGCATGCTCGTCACCGGTACCTTTGTCGGGTACCTCATCATATTCGCTGGTGCGGCCGCGGGCTACATAATGCAGACTCCTTCACACAAACGGATCGACATCTTCTATTCGCTGGTCGGTGTTGCCTTGTTCGTCGCTAGCGGTGCCATTATTATTGACAGATTCCAACATTATGGTAAGAGCGAGATCAAAGACAAGAACTTGGCTAAGGCCTCGTTGGCCATCATCAATGGCGCTATCTTACTTGTTGATGCTGTCCTCACGCAACGAGGAGGCTAA
Protein
MAISRLSIIKFLELALTCSCVALHYHSYNADADIGMLVTGTFVGYLIIFAGAAAGYIMQTPSHKRIDIFYSLVGVALFVASGAIIIDRFQHYGKSEIKDKNLAKASLAIINGAILLVDAVLTQRGG
Summary
Uniprot
A0FDQ7
A0A212F4M6
S4PET8
A0A1E1W631
A0A194PJZ6
A0A0L7L8Q0
+ More
A0A2A4IV10 A0A3S2P3T2 I4DNV8 A0A1Y1NIF3 T1DJ21 A0A3L8DHR6 A0A1W4XS59 T1E390 A0A158NKQ9 J3JVZ5 D6WWS3 C6ZQY5 F4WXC8 A0A1Q3FNB7 B0X9Z7 U4UA06 A0A023EFJ8 A0A182J558 A0A023EDL0 Q16SL0 U5ESI9 B4M2R4 B4NCR8 B4JJQ0 A0A182W454 B4L4E0 A0A182MZW2 A0A182UCP7 A0A182XNI5 Q7QI12 A0A182IDN3 Q29FV3 B4GWP2 A0A182RH69 B6DE37 A0A1W4VWR3 B4R3C9 B4PYL7 B3N0J3 Q9VRD7 A0A2M3YYI0 A0A2M4AAY7 W5JCW4 A0A182FIN9 A0A1J1IYD6 B3NY60 A0A182LD44 E0VCQ7 A0A2M4AKU3 A0A2M4C4B7 B6DE56 A0A182Q2Q8 A0A182VNW3 T1DPT9 A0A3B0JZA0 B0X9Z8 Q7PSX2 Q4QPX9 A0A182PW46 A0A0L0BPW3 Q16SK9 A0A1B0FPY3 A0A1A9XSL3 A0A1A9UEI0 A0A1B0AQG2 A0A1B0ADP1 A0A1S4GAK6 A0A067QM99 A0A182XNI6 A0A1B0CF78 A0A182S9X8 A0A1B0CST7 A0A182MZW1 A0A182QPP5 A0A0K8VBQ9 A0A026VZY4 A0A1S4FR32 A0A182RH70 A0A1B0DDX9 A0A182FM75 A0A1I8P197 A0A182JEN2 A0A195F682 E2ANQ1 A0A1I8MDV5 A0A2S2NP50 A0A195EJ94 A0A084VN04 E2BQR4 A0A232F4M4 A0A1L8D9L7
A0A2A4IV10 A0A3S2P3T2 I4DNV8 A0A1Y1NIF3 T1DJ21 A0A3L8DHR6 A0A1W4XS59 T1E390 A0A158NKQ9 J3JVZ5 D6WWS3 C6ZQY5 F4WXC8 A0A1Q3FNB7 B0X9Z7 U4UA06 A0A023EFJ8 A0A182J558 A0A023EDL0 Q16SL0 U5ESI9 B4M2R4 B4NCR8 B4JJQ0 A0A182W454 B4L4E0 A0A182MZW2 A0A182UCP7 A0A182XNI5 Q7QI12 A0A182IDN3 Q29FV3 B4GWP2 A0A182RH69 B6DE37 A0A1W4VWR3 B4R3C9 B4PYL7 B3N0J3 Q9VRD7 A0A2M3YYI0 A0A2M4AAY7 W5JCW4 A0A182FIN9 A0A1J1IYD6 B3NY60 A0A182LD44 E0VCQ7 A0A2M4AKU3 A0A2M4C4B7 B6DE56 A0A182Q2Q8 A0A182VNW3 T1DPT9 A0A3B0JZA0 B0X9Z8 Q7PSX2 Q4QPX9 A0A182PW46 A0A0L0BPW3 Q16SK9 A0A1B0FPY3 A0A1A9XSL3 A0A1A9UEI0 A0A1B0AQG2 A0A1B0ADP1 A0A1S4GAK6 A0A067QM99 A0A182XNI6 A0A1B0CF78 A0A182S9X8 A0A1B0CST7 A0A182MZW1 A0A182QPP5 A0A0K8VBQ9 A0A026VZY4 A0A1S4FR32 A0A182RH70 A0A1B0DDX9 A0A182FM75 A0A1I8P197 A0A182JEN2 A0A195F682 E2ANQ1 A0A1I8MDV5 A0A2S2NP50 A0A195EJ94 A0A084VN04 E2BQR4 A0A232F4M4 A0A1L8D9L7
Pubmed
22118469
23622113
26354079
26227816
22651552
28004739
+ More
24330624 30249741 21347285 22516182 18362917 19820115 20496585 21719571 23537049 24945155 17510324 17994087 12364791 14747013 17210077 15632085 19178717 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20920257 23761445 20966253 20566863 26108605 24845553 24508170 20798317 25315136 24438588 28648823
24330624 30249741 21347285 22516182 18362917 19820115 20496585 21719571 23537049 24945155 17510324 17994087 12364791 14747013 17210077 15632085 19178717 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20920257 23761445 20966253 20566863 26108605 24845553 24508170 20798317 25315136 24438588 28648823
EMBL
DQ985598
ABJ97187.1
AGBW02010343
OWR48679.1
GAIX01006905
JAA85655.1
+ More
GDQN01008612 JAT82442.1 KQ459603 KPI93049.1 JTDY01002190 KOB71913.1 NWSH01007415 NWSH01003836 PCG62980.1 PCG65777.1 RSAL01000031 RVE51653.1 AK403162 BAM19598.1 GEZM01003645 JAV96660.1 GALA01000576 JAA94276.1 QOIP01000008 RLU19388.1 GALA01000580 JAA94272.1 ADTU01002122 BT127413 AEE62375.1 KQ971361 EFA08752.1 EZ114936 ACU30989.1 GL888424 EGI61094.1 GFDL01005946 JAV29099.1 DS232556 EDS43423.1 KB632155 ERL89188.1 GAPW01006023 GAPW01006022 GAPW01006020 JAC07575.1 GAPW01006025 GAPW01006024 GAPW01006021 JAC07573.1 CH477672 EAT37454.1 GANO01004816 JAB55055.1 CH940651 EDW65968.1 CH964239 EDW82627.1 CH916370 EDV99802.1 CH933810 EDW07418.1 AAAB01008811 EAA04904.4 APCN01004748 CH379065 EAL31476.1 CH479194 EDW27126.1 EU934406 ACI30184.1 CM000366 EDX18500.1 CM000162 EDX03058.1 CH902640 EDV38397.1 AE014298 KX531666 AAF50865.1 ANY27476.1 GGFM01000578 MBW21329.1 GGFK01004630 MBW37951.1 ADMH02001869 ETN60730.1 CVRI01000063 CRL04714.1 CH954180 EDV47581.1 DS235061 EEB11163.1 GGFK01008088 MBW41409.1 GGFJ01010697 MBW59838.1 EU934425 ACI30203.1 AXCN02002024 GAMD01003049 JAA98541.1 OUUW01000003 SPP78051.1 EDS43424.1 EAA04905.4 BT023637 AAY85037.1 JRES01001642 KNC21259.1 EAT37455.1 CCAG010020168 JXJN01001851 KK853177 KDR10266.1 AJWK01009788 AJWK01026568 GDHF01015915 JAI36399.1 KK107536 EZA49205.1 AJVK01005529 KQ981805 KYN35574.1 GL441295 EFN64937.1 GGMR01006354 MBY18973.1 KQ978801 KYN28323.1 ATLV01014663 KE524978 KFB39348.1 GL449798 EFN81983.1 NNAY01000947 OXU25786.1 GFDF01010925 JAV03159.1
GDQN01008612 JAT82442.1 KQ459603 KPI93049.1 JTDY01002190 KOB71913.1 NWSH01007415 NWSH01003836 PCG62980.1 PCG65777.1 RSAL01000031 RVE51653.1 AK403162 BAM19598.1 GEZM01003645 JAV96660.1 GALA01000576 JAA94276.1 QOIP01000008 RLU19388.1 GALA01000580 JAA94272.1 ADTU01002122 BT127413 AEE62375.1 KQ971361 EFA08752.1 EZ114936 ACU30989.1 GL888424 EGI61094.1 GFDL01005946 JAV29099.1 DS232556 EDS43423.1 KB632155 ERL89188.1 GAPW01006023 GAPW01006022 GAPW01006020 JAC07575.1 GAPW01006025 GAPW01006024 GAPW01006021 JAC07573.1 CH477672 EAT37454.1 GANO01004816 JAB55055.1 CH940651 EDW65968.1 CH964239 EDW82627.1 CH916370 EDV99802.1 CH933810 EDW07418.1 AAAB01008811 EAA04904.4 APCN01004748 CH379065 EAL31476.1 CH479194 EDW27126.1 EU934406 ACI30184.1 CM000366 EDX18500.1 CM000162 EDX03058.1 CH902640 EDV38397.1 AE014298 KX531666 AAF50865.1 ANY27476.1 GGFM01000578 MBW21329.1 GGFK01004630 MBW37951.1 ADMH02001869 ETN60730.1 CVRI01000063 CRL04714.1 CH954180 EDV47581.1 DS235061 EEB11163.1 GGFK01008088 MBW41409.1 GGFJ01010697 MBW59838.1 EU934425 ACI30203.1 AXCN02002024 GAMD01003049 JAA98541.1 OUUW01000003 SPP78051.1 EDS43424.1 EAA04905.4 BT023637 AAY85037.1 JRES01001642 KNC21259.1 EAT37455.1 CCAG010020168 JXJN01001851 KK853177 KDR10266.1 AJWK01009788 AJWK01026568 GDHF01015915 JAI36399.1 KK107536 EZA49205.1 AJVK01005529 KQ981805 KYN35574.1 GL441295 EFN64937.1 GGMR01006354 MBY18973.1 KQ978801 KYN28323.1 ATLV01014663 KE524978 KFB39348.1 GL449798 EFN81983.1 NNAY01000947 OXU25786.1 GFDF01010925 JAV03159.1
Proteomes
UP000007151
UP000053268
UP000037510
UP000218220
UP000283053
UP000279307
+ More
UP000192223 UP000005205 UP000007266 UP000007755 UP000002320 UP000030742 UP000075880 UP000008820 UP000008792 UP000007798 UP000001070 UP000075920 UP000009192 UP000075884 UP000075902 UP000076407 UP000007062 UP000075840 UP000001819 UP000008744 UP000075900 UP000192221 UP000000304 UP000002282 UP000007801 UP000000803 UP000000673 UP000069272 UP000183832 UP000008711 UP000075882 UP000009046 UP000075886 UP000075903 UP000268350 UP000075885 UP000037069 UP000092444 UP000092443 UP000078200 UP000092460 UP000092445 UP000027135 UP000092461 UP000075901 UP000053097 UP000092462 UP000095300 UP000078541 UP000000311 UP000095301 UP000078492 UP000030765 UP000008237 UP000215335
UP000192223 UP000005205 UP000007266 UP000007755 UP000002320 UP000030742 UP000075880 UP000008820 UP000008792 UP000007798 UP000001070 UP000075920 UP000009192 UP000075884 UP000075902 UP000076407 UP000007062 UP000075840 UP000001819 UP000008744 UP000075900 UP000192221 UP000000304 UP000002282 UP000007801 UP000000803 UP000000673 UP000069272 UP000183832 UP000008711 UP000075882 UP000009046 UP000075886 UP000075903 UP000268350 UP000075885 UP000037069 UP000092444 UP000092443 UP000078200 UP000092460 UP000092445 UP000027135 UP000092461 UP000075901 UP000053097 UP000092462 UP000095300 UP000078541 UP000000311 UP000095301 UP000078492 UP000030765 UP000008237 UP000215335
Interpro
IPR038976
Ssk
ProteinModelPortal
A0FDQ7
A0A212F4M6
S4PET8
A0A1E1W631
A0A194PJZ6
A0A0L7L8Q0
+ More
A0A2A4IV10 A0A3S2P3T2 I4DNV8 A0A1Y1NIF3 T1DJ21 A0A3L8DHR6 A0A1W4XS59 T1E390 A0A158NKQ9 J3JVZ5 D6WWS3 C6ZQY5 F4WXC8 A0A1Q3FNB7 B0X9Z7 U4UA06 A0A023EFJ8 A0A182J558 A0A023EDL0 Q16SL0 U5ESI9 B4M2R4 B4NCR8 B4JJQ0 A0A182W454 B4L4E0 A0A182MZW2 A0A182UCP7 A0A182XNI5 Q7QI12 A0A182IDN3 Q29FV3 B4GWP2 A0A182RH69 B6DE37 A0A1W4VWR3 B4R3C9 B4PYL7 B3N0J3 Q9VRD7 A0A2M3YYI0 A0A2M4AAY7 W5JCW4 A0A182FIN9 A0A1J1IYD6 B3NY60 A0A182LD44 E0VCQ7 A0A2M4AKU3 A0A2M4C4B7 B6DE56 A0A182Q2Q8 A0A182VNW3 T1DPT9 A0A3B0JZA0 B0X9Z8 Q7PSX2 Q4QPX9 A0A182PW46 A0A0L0BPW3 Q16SK9 A0A1B0FPY3 A0A1A9XSL3 A0A1A9UEI0 A0A1B0AQG2 A0A1B0ADP1 A0A1S4GAK6 A0A067QM99 A0A182XNI6 A0A1B0CF78 A0A182S9X8 A0A1B0CST7 A0A182MZW1 A0A182QPP5 A0A0K8VBQ9 A0A026VZY4 A0A1S4FR32 A0A182RH70 A0A1B0DDX9 A0A182FM75 A0A1I8P197 A0A182JEN2 A0A195F682 E2ANQ1 A0A1I8MDV5 A0A2S2NP50 A0A195EJ94 A0A084VN04 E2BQR4 A0A232F4M4 A0A1L8D9L7
A0A2A4IV10 A0A3S2P3T2 I4DNV8 A0A1Y1NIF3 T1DJ21 A0A3L8DHR6 A0A1W4XS59 T1E390 A0A158NKQ9 J3JVZ5 D6WWS3 C6ZQY5 F4WXC8 A0A1Q3FNB7 B0X9Z7 U4UA06 A0A023EFJ8 A0A182J558 A0A023EDL0 Q16SL0 U5ESI9 B4M2R4 B4NCR8 B4JJQ0 A0A182W454 B4L4E0 A0A182MZW2 A0A182UCP7 A0A182XNI5 Q7QI12 A0A182IDN3 Q29FV3 B4GWP2 A0A182RH69 B6DE37 A0A1W4VWR3 B4R3C9 B4PYL7 B3N0J3 Q9VRD7 A0A2M3YYI0 A0A2M4AAY7 W5JCW4 A0A182FIN9 A0A1J1IYD6 B3NY60 A0A182LD44 E0VCQ7 A0A2M4AKU3 A0A2M4C4B7 B6DE56 A0A182Q2Q8 A0A182VNW3 T1DPT9 A0A3B0JZA0 B0X9Z8 Q7PSX2 Q4QPX9 A0A182PW46 A0A0L0BPW3 Q16SK9 A0A1B0FPY3 A0A1A9XSL3 A0A1A9UEI0 A0A1B0AQG2 A0A1B0ADP1 A0A1S4GAK6 A0A067QM99 A0A182XNI6 A0A1B0CF78 A0A182S9X8 A0A1B0CST7 A0A182MZW1 A0A182QPP5 A0A0K8VBQ9 A0A026VZY4 A0A1S4FR32 A0A182RH70 A0A1B0DDX9 A0A182FM75 A0A1I8P197 A0A182JEN2 A0A195F682 E2ANQ1 A0A1I8MDV5 A0A2S2NP50 A0A195EJ94 A0A084VN04 E2BQR4 A0A232F4M4 A0A1L8D9L7
Ontologies
PANTHER
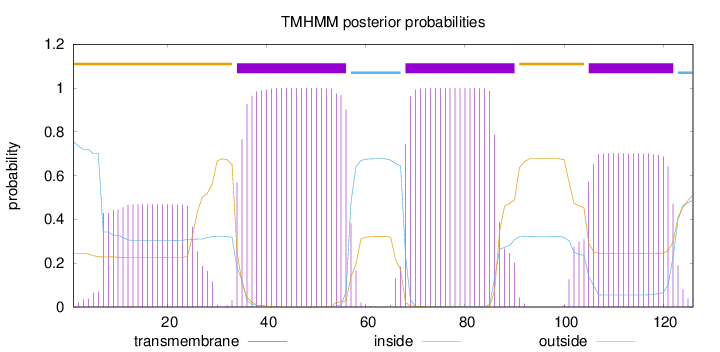
Topology
Length:
126
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
65.66349
Exp number, first 60 AAs:
32.25023
Total prob of N-in:
0.75549
POSSIBLE N-term signal
sequence
outside
1 - 33
TMhelix
34 - 56
inside
57 - 67
TMhelix
68 - 90
outside
91 - 104
TMhelix
105 - 122
inside
123 - 126
Population Genetic Test Statistics
Pi
192.707424
Theta
162.128524
Tajima's D
0.43273
CLR
0
CSRT
0.495125243737813
Interpretation
Uncertain
