Gene
KWMTBOMO10427
Pre Gene Modal
BGIBMGA007019
Annotation
putative_mitochondrial_ribosomal_protein_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.999
Sequence
CDS
ATGGCCAGTAGCAGATTGGAAAGAATTGGTACAATATTTACCAGAGTTGAAGGTCTTCTTAGCCGAGGAGCAATGAAACCTGATGACAGACCCTTGTGGTTTGATGTATACAAAGCTTTCCCTCCAATCACCGAACCTAAATATGCACGTCCTAATCTTGTAGTCAAAGAAATACGACCAATCTTATACAAGGAAGACGTTTTAAGAGCAAAATTCCATTCAAACGGTTACGGTTTAGCACCAGTAAGCCTATTAAATCAAAGTAATGAAACTCAAACAAAACGTCTTGTACAGCAATACGATGAATTAAAAGCAGAAGGCATTCCTGAAGATGAAATCATTGAGAAAGCCGCACAAGCAGTAGCCGTCGAACGCCACAGCTATGCTGCACAAAAATTGAATGTTACACCTAAAAATCCTGATTCCGTTACAGCCCAAGTATTGGCTGAAGCTGATATAAAAAATATTTTCAACAACAAATAA
Protein
MASSRLERIGTIFTRVEGLLSRGAMKPDDRPLWFDVYKAFPPITEPKYARPNLVVKEIRPILYKEDVLRAKFHSNGYGLAPVSLLNQSNETQTKRLVQQYDELKAEGIPEDEIIEKAAQAVAVERHSYAAQKLNVTPKNPDSVTAQVLAEADIKNIFNNK
Summary
Uniprot
A0FDR0
H9JBX4
A0A194QUY3
A0A0L7L870
A0A1E1WSS3
A0A194PJY6
+ More
A0A212FB60 A0A1A9X0R9 B4MVQ8 A0A1I8N6U7 A0A1A9V9J2 A0A1B0A537 B4HXA8 A0A3B0KBU5 A0A1B0DRG0 A0A1I8Q705 B4Q574 B4KEN6 B4JCY0 A0A067R5F4 A0A1W4VKE1 A0A182MDZ6 A0A034WQZ9 A0A0C9R5R6 A0A182F982 A0A336LMB5 Q8IP62 A0A1L8E001 B3NAC5 A0A1A9XL11 A0A1L8E095 A0A182LYQ8 Q7QD03 A0A182II33 A0A182LGA0 B4P3R1 A0A0K8TVQ1 A0A182UUG0 A0A182QUL5 Q29K79 A0A2M4ASR6 A0A0M4E927 A0A0P4WKZ3 B4GX17 B3MN17 B4M8M0 A0A0C9RUB6 A0A182XW93 A0A182N705 A0A0P5VPZ8 A0A023EHY9 A0A0P5FNG0 A0A224XZY4 A0A2M3ZJ88 A0A2M4C1R9 A0A2M3ZJA2 A0A1B0G935 A0A2M4C1Q9 W5JSP7 A0A2M3ZCH7 A0A182TB17 A0A1B1SN29 A0A084VG89 A0A0P5BKI0 A0A2M4C1P4 A0A182XHJ3 A0A1S4FC32 W8BSU6 U5EFL6 A0A0V0GBW3 A0A182J4G8 A0A1Q3FX00 Q178C3 A0A182HA90 E9G259 A0A3S3P719 A0A3B5JZ70 A0A182VYP1 T1IQ79 A0A2S2QY75 D6WUN6 A0A2R5LHL3 A0A026WY45 A0A293N362 A0A1W4WX76 A0A1W0X4C7 A0A151XBX2 A0A0P4VPY1 H3B5A1 A0A1B6G1M3 A0A1Z5L7Z0 A0A1Y1LFJ9 R7TAJ1 K7IZG9 A0A1B6JL41
A0A212FB60 A0A1A9X0R9 B4MVQ8 A0A1I8N6U7 A0A1A9V9J2 A0A1B0A537 B4HXA8 A0A3B0KBU5 A0A1B0DRG0 A0A1I8Q705 B4Q574 B4KEN6 B4JCY0 A0A067R5F4 A0A1W4VKE1 A0A182MDZ6 A0A034WQZ9 A0A0C9R5R6 A0A182F982 A0A336LMB5 Q8IP62 A0A1L8E001 B3NAC5 A0A1A9XL11 A0A1L8E095 A0A182LYQ8 Q7QD03 A0A182II33 A0A182LGA0 B4P3R1 A0A0K8TVQ1 A0A182UUG0 A0A182QUL5 Q29K79 A0A2M4ASR6 A0A0M4E927 A0A0P4WKZ3 B4GX17 B3MN17 B4M8M0 A0A0C9RUB6 A0A182XW93 A0A182N705 A0A0P5VPZ8 A0A023EHY9 A0A0P5FNG0 A0A224XZY4 A0A2M3ZJ88 A0A2M4C1R9 A0A2M3ZJA2 A0A1B0G935 A0A2M4C1Q9 W5JSP7 A0A2M3ZCH7 A0A182TB17 A0A1B1SN29 A0A084VG89 A0A0P5BKI0 A0A2M4C1P4 A0A182XHJ3 A0A1S4FC32 W8BSU6 U5EFL6 A0A0V0GBW3 A0A182J4G8 A0A1Q3FX00 Q178C3 A0A182HA90 E9G259 A0A3S3P719 A0A3B5JZ70 A0A182VYP1 T1IQ79 A0A2S2QY75 D6WUN6 A0A2R5LHL3 A0A026WY45 A0A293N362 A0A1W4WX76 A0A1W0X4C7 A0A151XBX2 A0A0P4VPY1 H3B5A1 A0A1B6G1M3 A0A1Z5L7Z0 A0A1Y1LFJ9 R7TAJ1 K7IZG9 A0A1B6JL41
Pubmed
19121390
26354079
26227816
22118469
17994087
25315136
+ More
22936249 24845553 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12364791 14747013 17210077 20966253 17550304 15632085 25244985 24945155 20920257 23761445 24438588 24495485 17510324 26483478 21292972 21551351 18362917 19820115 24508170 27129103 9215903 28528879 28004739 23254933 20075255
22936249 24845553 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12364791 14747013 17210077 20966253 17550304 15632085 25244985 24945155 20920257 23761445 24438588 24495485 17510324 26483478 21292972 21551351 18362917 19820115 24508170 27129103 9215903 28528879 28004739 23254933 20075255
EMBL
DQ985601
ABJ97190.1
BABH01014833
KQ461137
KPJ08790.1
JTDY01002381
+ More
KOB71534.1 GDQN01001004 JAT90050.1 KQ459603 KPI93039.1 AGBW02009379 OWR50959.1 CH963857 EDW75778.1 CH480818 EDW51688.1 OUUW01000006 SPP82531.1 AJVK01019990 CM000361 CM002910 EDX04980.1 KMY90173.1 CH933807 EDW11915.1 CH916368 EDW03219.1 KK852687 KDR18398.1 AXCM01006850 GAKP01002337 JAC56615.1 GBYB01003260 JAG73027.1 UFQS01002906 UFQT01002906 SSX14862.1 SSX34250.1 AE014134 AAN10854.1 GFDF01001994 JAV12090.1 CH954177 EDV58627.1 GFDF01001995 JAV12089.1 AXCM01009416 AAAB01008859 EAA08013.5 APCN01000586 CM000157 EDW88362.1 GDHF01033956 JAI18358.1 AXCN02000321 CH379061 EAL33298.1 GGFK01010500 MBW43821.1 CP012523 ALC39540.1 GDRN01010187 JAI67916.1 CH479195 EDW27344.1 CH902620 EDV31995.1 CH940654 EDW57546.1 GBYB01012395 JAG82162.1 GDIP01097067 LRGB01002384 JAM06648.1 KZS07902.1 GAPW01005092 JAC08506.1 GDIQ01258685 JAJ93039.1 GFTR01002573 JAW13853.1 GGFM01007896 MBW28647.1 GGFJ01010104 MBW59245.1 GGFM01007863 MBW28614.1 CCAG010004816 GGFJ01010102 MBW59243.1 ADMH02000272 ETN67166.1 GGFM01005460 MBW26211.1 KU513760 ANV22166.1 ATLV01012704 KE524809 KFB36983.1 GDIP01187456 JAJ35946.1 GGFJ01010105 MBW59246.1 GAMC01004328 JAC02228.1 GANO01003791 JAB56080.1 GECL01001287 JAP04837.1 GFDL01002911 JAV32134.1 CH477366 EAT42530.1 JXUM01030874 KQ560899 KXJ80432.1 GL732530 EFX86364.1 NCKU01000258 RWS16273.1 JH431295 GGMS01013247 MBY82450.1 KQ971363 EFA07822.2 GGLE01004812 MBY08938.1 KK107063 EZA60995.1 GFWV01022766 MAA47493.1 MTYJ01000018 OQV22329.1 KQ982320 KYQ57810.1 GDKW01002058 JAI54537.1 AFYH01008348 AFYH01008349 GECZ01013434 JAS56335.1 GFJQ02003816 JAW03154.1 GEZM01058821 GEZM01058820 JAV71658.1 AMQN01014198 AMQN01014199 KB310797 ELT90724.1 GECU01007758 JAS99948.1
KOB71534.1 GDQN01001004 JAT90050.1 KQ459603 KPI93039.1 AGBW02009379 OWR50959.1 CH963857 EDW75778.1 CH480818 EDW51688.1 OUUW01000006 SPP82531.1 AJVK01019990 CM000361 CM002910 EDX04980.1 KMY90173.1 CH933807 EDW11915.1 CH916368 EDW03219.1 KK852687 KDR18398.1 AXCM01006850 GAKP01002337 JAC56615.1 GBYB01003260 JAG73027.1 UFQS01002906 UFQT01002906 SSX14862.1 SSX34250.1 AE014134 AAN10854.1 GFDF01001994 JAV12090.1 CH954177 EDV58627.1 GFDF01001995 JAV12089.1 AXCM01009416 AAAB01008859 EAA08013.5 APCN01000586 CM000157 EDW88362.1 GDHF01033956 JAI18358.1 AXCN02000321 CH379061 EAL33298.1 GGFK01010500 MBW43821.1 CP012523 ALC39540.1 GDRN01010187 JAI67916.1 CH479195 EDW27344.1 CH902620 EDV31995.1 CH940654 EDW57546.1 GBYB01012395 JAG82162.1 GDIP01097067 LRGB01002384 JAM06648.1 KZS07902.1 GAPW01005092 JAC08506.1 GDIQ01258685 JAJ93039.1 GFTR01002573 JAW13853.1 GGFM01007896 MBW28647.1 GGFJ01010104 MBW59245.1 GGFM01007863 MBW28614.1 CCAG010004816 GGFJ01010102 MBW59243.1 ADMH02000272 ETN67166.1 GGFM01005460 MBW26211.1 KU513760 ANV22166.1 ATLV01012704 KE524809 KFB36983.1 GDIP01187456 JAJ35946.1 GGFJ01010105 MBW59246.1 GAMC01004328 JAC02228.1 GANO01003791 JAB56080.1 GECL01001287 JAP04837.1 GFDL01002911 JAV32134.1 CH477366 EAT42530.1 JXUM01030874 KQ560899 KXJ80432.1 GL732530 EFX86364.1 NCKU01000258 RWS16273.1 JH431295 GGMS01013247 MBY82450.1 KQ971363 EFA07822.2 GGLE01004812 MBY08938.1 KK107063 EZA60995.1 GFWV01022766 MAA47493.1 MTYJ01000018 OQV22329.1 KQ982320 KYQ57810.1 GDKW01002058 JAI54537.1 AFYH01008348 AFYH01008349 GECZ01013434 JAS56335.1 GFJQ02003816 JAW03154.1 GEZM01058821 GEZM01058820 JAV71658.1 AMQN01014198 AMQN01014199 KB310797 ELT90724.1 GECU01007758 JAS99948.1
Proteomes
UP000005204
UP000053240
UP000037510
UP000053268
UP000007151
UP000091820
+ More
UP000007798 UP000095301 UP000078200 UP000092445 UP000001292 UP000268350 UP000092462 UP000095300 UP000000304 UP000009192 UP000001070 UP000027135 UP000192221 UP000075883 UP000069272 UP000000803 UP000008711 UP000092443 UP000007062 UP000075840 UP000075882 UP000002282 UP000075903 UP000075886 UP000001819 UP000092553 UP000008744 UP000007801 UP000008792 UP000076408 UP000075884 UP000076858 UP000092444 UP000000673 UP000075901 UP000030765 UP000076407 UP000075880 UP000008820 UP000069940 UP000249989 UP000000305 UP000285301 UP000005226 UP000075920 UP000007266 UP000053097 UP000192223 UP000075809 UP000008672 UP000014760 UP000002358
UP000007798 UP000095301 UP000078200 UP000092445 UP000001292 UP000268350 UP000092462 UP000095300 UP000000304 UP000009192 UP000001070 UP000027135 UP000192221 UP000075883 UP000069272 UP000000803 UP000008711 UP000092443 UP000007062 UP000075840 UP000075882 UP000002282 UP000075903 UP000075886 UP000001819 UP000092553 UP000008744 UP000007801 UP000008792 UP000076408 UP000075884 UP000076858 UP000092444 UP000000673 UP000075901 UP000030765 UP000076407 UP000075880 UP000008820 UP000069940 UP000249989 UP000000305 UP000285301 UP000005226 UP000075920 UP000007266 UP000053097 UP000192223 UP000075809 UP000008672 UP000014760 UP000002358
Pfam
Interpro
IPR023611
Ribosomal_S23/S25_mit
+ More
IPR019520 Ribosomal_S23
IPR014772 Munc13_dom-2
IPR006888 XLR/SYCP3/FAM9_dom
IPR000742 EGF-like_dom
IPR013032 EGF-like_CS
IPR002809 EMC3/TMCO1
IPR019333 Int_cplx_su3
IPR011004 Trimer_LpxA-like_sf
IPR002049 Laminin_EGF
IPR011489 EMI_domain
IPR001451 Hexapep
IPR035979 RBD_domain_sf
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR000504 RRM_dom
IPR019520 Ribosomal_S23
IPR014772 Munc13_dom-2
IPR006888 XLR/SYCP3/FAM9_dom
IPR000742 EGF-like_dom
IPR013032 EGF-like_CS
IPR002809 EMC3/TMCO1
IPR019333 Int_cplx_su3
IPR011004 Trimer_LpxA-like_sf
IPR002049 Laminin_EGF
IPR011489 EMI_domain
IPR001451 Hexapep
IPR035979 RBD_domain_sf
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR000504 RRM_dom
Gene 3D
ProteinModelPortal
A0FDR0
H9JBX4
A0A194QUY3
A0A0L7L870
A0A1E1WSS3
A0A194PJY6
+ More
A0A212FB60 A0A1A9X0R9 B4MVQ8 A0A1I8N6U7 A0A1A9V9J2 A0A1B0A537 B4HXA8 A0A3B0KBU5 A0A1B0DRG0 A0A1I8Q705 B4Q574 B4KEN6 B4JCY0 A0A067R5F4 A0A1W4VKE1 A0A182MDZ6 A0A034WQZ9 A0A0C9R5R6 A0A182F982 A0A336LMB5 Q8IP62 A0A1L8E001 B3NAC5 A0A1A9XL11 A0A1L8E095 A0A182LYQ8 Q7QD03 A0A182II33 A0A182LGA0 B4P3R1 A0A0K8TVQ1 A0A182UUG0 A0A182QUL5 Q29K79 A0A2M4ASR6 A0A0M4E927 A0A0P4WKZ3 B4GX17 B3MN17 B4M8M0 A0A0C9RUB6 A0A182XW93 A0A182N705 A0A0P5VPZ8 A0A023EHY9 A0A0P5FNG0 A0A224XZY4 A0A2M3ZJ88 A0A2M4C1R9 A0A2M3ZJA2 A0A1B0G935 A0A2M4C1Q9 W5JSP7 A0A2M3ZCH7 A0A182TB17 A0A1B1SN29 A0A084VG89 A0A0P5BKI0 A0A2M4C1P4 A0A182XHJ3 A0A1S4FC32 W8BSU6 U5EFL6 A0A0V0GBW3 A0A182J4G8 A0A1Q3FX00 Q178C3 A0A182HA90 E9G259 A0A3S3P719 A0A3B5JZ70 A0A182VYP1 T1IQ79 A0A2S2QY75 D6WUN6 A0A2R5LHL3 A0A026WY45 A0A293N362 A0A1W4WX76 A0A1W0X4C7 A0A151XBX2 A0A0P4VPY1 H3B5A1 A0A1B6G1M3 A0A1Z5L7Z0 A0A1Y1LFJ9 R7TAJ1 K7IZG9 A0A1B6JL41
A0A212FB60 A0A1A9X0R9 B4MVQ8 A0A1I8N6U7 A0A1A9V9J2 A0A1B0A537 B4HXA8 A0A3B0KBU5 A0A1B0DRG0 A0A1I8Q705 B4Q574 B4KEN6 B4JCY0 A0A067R5F4 A0A1W4VKE1 A0A182MDZ6 A0A034WQZ9 A0A0C9R5R6 A0A182F982 A0A336LMB5 Q8IP62 A0A1L8E001 B3NAC5 A0A1A9XL11 A0A1L8E095 A0A182LYQ8 Q7QD03 A0A182II33 A0A182LGA0 B4P3R1 A0A0K8TVQ1 A0A182UUG0 A0A182QUL5 Q29K79 A0A2M4ASR6 A0A0M4E927 A0A0P4WKZ3 B4GX17 B3MN17 B4M8M0 A0A0C9RUB6 A0A182XW93 A0A182N705 A0A0P5VPZ8 A0A023EHY9 A0A0P5FNG0 A0A224XZY4 A0A2M3ZJ88 A0A2M4C1R9 A0A2M3ZJA2 A0A1B0G935 A0A2M4C1Q9 W5JSP7 A0A2M3ZCH7 A0A182TB17 A0A1B1SN29 A0A084VG89 A0A0P5BKI0 A0A2M4C1P4 A0A182XHJ3 A0A1S4FC32 W8BSU6 U5EFL6 A0A0V0GBW3 A0A182J4G8 A0A1Q3FX00 Q178C3 A0A182HA90 E9G259 A0A3S3P719 A0A3B5JZ70 A0A182VYP1 T1IQ79 A0A2S2QY75 D6WUN6 A0A2R5LHL3 A0A026WY45 A0A293N362 A0A1W4WX76 A0A1W0X4C7 A0A151XBX2 A0A0P4VPY1 H3B5A1 A0A1B6G1M3 A0A1Z5L7Z0 A0A1Y1LFJ9 R7TAJ1 K7IZG9 A0A1B6JL41
PDB
6NU3
E-value=4.60697e-17,
Score=209
Ontologies
KEGG
GO
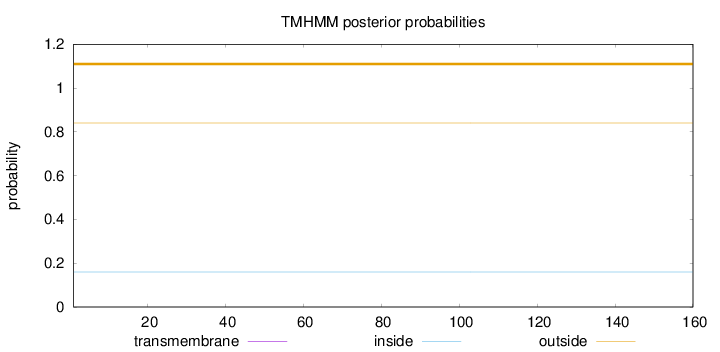
Topology
Length:
160
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00032
Exp number, first 60 AAs:
0
Total prob of N-in:
0.15954
outside
1 - 160
Population Genetic Test Statistics
Pi
134.310365
Theta
156.314474
Tajima's D
-0.697139
CLR
0.195293
CSRT
0.197090145492725
Interpretation
Uncertain
