Gene
KWMTBOMO10425
Annotation
PREDICTED:_uncharacterized_protein_LOC105842480_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.635
Sequence
CDS
ATGGCACAGAAGAAATCTGTTTTGTTTTACTTCGCTGTGATTTTAGTTATTTTGAGTACACTTACACAAGAGGTAGAAGCCAGAAGAAAAATTCTTAGAGGAAGACGTGTTATGACCCGCACTTACTACCGTGGCAATGCGGTTCCCGCGTGGGCAATTAGCTTGATGGCTGGAATTGGAATGTTGATCATCGGTGGCGTCTTATATGTAGTTATGAGGAAACTCGTTCTATCTTCGGAAACGGGTTCACTGAATACTTACCAACCTGCTATGCAGCAAGAAGTATAA
Protein
MAQKKSVLFYFAVILVILSTLTQEVEARRKILRGRRVMTRTYYRGNAVPAWAISLMAGIGMLIIGGVLYVVMRKLVLSSETGSLNTYQPAMQQEV
Summary
Uniprot
A0A194QTZ8
A0A194PJ13
A0A3S2LR59
Q16ZV7
B0WXN7
A0A023ECJ5
+ More
A0A1Q3FL49 A0A1Q3FKU0 A0A182QWW1 A0A182R6C8 Q7PYM4 A0A182VZ14 A0A182V5Y7 W5JJ86 T1EAP0 A0A154PSP8 A0A0L7R2Y7 A0A182JK68 U5ED05 A0A336M033 A0A1B0D512 A0A1Y1LH00 A0A3L8DD20 E2C4G1 A0A1I8M343 K7JGT6 A0A232ERU0 A0A067QXH5 A0A1J1IG39 B4MMW4 A0A088A5M0 A0A1L8EB14 A0A084VHD4 A0A182PKK7 A0A1L8EG28 A0A158NFG7 A0A1I8PJ61 A0A1W4W405 A0A310SLD9 F5HM97 A0A0J7KJF3 A0A182XVX1 E9ITG7 A0A182I057 A0A139WLL4 A0A182FKT1 A0A182XBB4 A0A182NCF2 A0A182JWP4 B4IZ57 E2A6R5 B4MG12 B4KXC6 A0A195BDK4 Q29E95 B4HD20 A0A3B0JV09 A0A2A3E3M4 B3M9L5 B3NG76 A0A151WPE6 A0A224XWP0 F4WGU0 A0A1W4VLR6 A0A151JVB0 B4PIC7 T1HX73 A0A1B6H4G4 Q8SXS4 B4QR38 B4HUC0 A0A195DC25 A0A0L0BM80 A0A1L8DAF8 A0A0M8ZQ93 A0A0M4EFL2 A0A1S3DFQ1 A0A1A9ZUE8 A0A1L8D9X0 A0A1L8D9L1 A0A1B0FC04
A0A1Q3FL49 A0A1Q3FKU0 A0A182QWW1 A0A182R6C8 Q7PYM4 A0A182VZ14 A0A182V5Y7 W5JJ86 T1EAP0 A0A154PSP8 A0A0L7R2Y7 A0A182JK68 U5ED05 A0A336M033 A0A1B0D512 A0A1Y1LH00 A0A3L8DD20 E2C4G1 A0A1I8M343 K7JGT6 A0A232ERU0 A0A067QXH5 A0A1J1IG39 B4MMW4 A0A088A5M0 A0A1L8EB14 A0A084VHD4 A0A182PKK7 A0A1L8EG28 A0A158NFG7 A0A1I8PJ61 A0A1W4W405 A0A310SLD9 F5HM97 A0A0J7KJF3 A0A182XVX1 E9ITG7 A0A182I057 A0A139WLL4 A0A182FKT1 A0A182XBB4 A0A182NCF2 A0A182JWP4 B4IZ57 E2A6R5 B4MG12 B4KXC6 A0A195BDK4 Q29E95 B4HD20 A0A3B0JV09 A0A2A3E3M4 B3M9L5 B3NG76 A0A151WPE6 A0A224XWP0 F4WGU0 A0A1W4VLR6 A0A151JVB0 B4PIC7 T1HX73 A0A1B6H4G4 Q8SXS4 B4QR38 B4HUC0 A0A195DC25 A0A0L0BM80 A0A1L8DAF8 A0A0M8ZQ93 A0A0M4EFL2 A0A1S3DFQ1 A0A1A9ZUE8 A0A1L8D9X0 A0A1L8D9L1 A0A1B0FC04
Pubmed
26354079
17510324
24945155
26483478
12364791
14747013
+ More
17210077 20920257 23761445 28004739 30249741 20798317 25315136 20075255 28648823 24845553 17994087 24438588 21347285 25244985 21282665 18362917 19820115 15632085 21719571 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 26108605
17210077 20920257 23761445 28004739 30249741 20798317 25315136 20075255 28648823 24845553 17994087 24438588 21347285 25244985 21282665 18362917 19820115 15632085 21719571 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 26108605
EMBL
KQ461137
KPJ08789.1
KQ459603
KPI93038.1
RSAL01000374
RVE42126.1
+ More
CH477482 EAT40219.1 DS232169 EDS36567.1 JXUM01108704 GAPW01006535 KQ565257 JAC07063.1 KXJ71165.1 GFDL01006849 JAV28196.1 GFDL01006848 JAV28197.1 AXCN02000019 AAAB01008987 EAA01460.2 ADMH02001343 ETN62950.1 GAMD01000242 JAB01349.1 KQ435056 KZC14288.1 KQ414665 KOC65141.1 GANO01004917 JAB54954.1 UFQT01000367 SSX23592.1 AJVK01011686 GEZM01055874 JAV72903.1 QOIP01000010 RLU17789.1 GL452471 EFN77145.1 NNAY01002580 OXU21006.1 KK853134 KDR10842.1 CVRI01000049 CRK99184.1 CH963847 EDW73520.1 GFDG01003036 JAV15763.1 ATLV01013152 KE524842 KFB37378.1 GFDG01001119 JAV17680.1 ADTU01014479 KQ760116 OAD61944.1 EGK97419.1 LBMM01006736 KMQ90369.1 GL765555 EFZ16109.1 APCN01002045 KQ971321 KYB28796.1 CH916366 EDV95579.1 GL437197 EFN70859.1 CH940669 EDW58273.1 CH933809 EDW17584.1 KQ976514 KYM82265.1 CH379070 EAL30166.2 CH479611 EDW35121.1 OUUW01000002 SPP77549.1 KZ288400 PBC26288.1 CH902618 EDV39021.1 CH954178 EDV50977.1 KQ982877 KYQ49700.1 GFTR01000772 JAW15654.1 GL888147 EGI66559.1 KQ981724 KYN37240.1 CM000159 EDW93469.1 ACPB03010999 GECZ01000211 JAS69558.1 AE014296 AY084166 KX531355 AAF47950.2 AAL89904.1 ANY27165.1 CM000363 CM002912 EDX09275.1 KMY97691.1 CH480817 EDW50541.1 KQ981063 KYN09959.1 JRES01001653 KNC21127.1 GFDF01010780 JAV03304.1 KQ435912 KOX68895.1 CP012525 ALC43065.1 GFDF01010927 JAV03157.1 GFDF01010926 JAV03158.1 CCAG010011675
CH477482 EAT40219.1 DS232169 EDS36567.1 JXUM01108704 GAPW01006535 KQ565257 JAC07063.1 KXJ71165.1 GFDL01006849 JAV28196.1 GFDL01006848 JAV28197.1 AXCN02000019 AAAB01008987 EAA01460.2 ADMH02001343 ETN62950.1 GAMD01000242 JAB01349.1 KQ435056 KZC14288.1 KQ414665 KOC65141.1 GANO01004917 JAB54954.1 UFQT01000367 SSX23592.1 AJVK01011686 GEZM01055874 JAV72903.1 QOIP01000010 RLU17789.1 GL452471 EFN77145.1 NNAY01002580 OXU21006.1 KK853134 KDR10842.1 CVRI01000049 CRK99184.1 CH963847 EDW73520.1 GFDG01003036 JAV15763.1 ATLV01013152 KE524842 KFB37378.1 GFDG01001119 JAV17680.1 ADTU01014479 KQ760116 OAD61944.1 EGK97419.1 LBMM01006736 KMQ90369.1 GL765555 EFZ16109.1 APCN01002045 KQ971321 KYB28796.1 CH916366 EDV95579.1 GL437197 EFN70859.1 CH940669 EDW58273.1 CH933809 EDW17584.1 KQ976514 KYM82265.1 CH379070 EAL30166.2 CH479611 EDW35121.1 OUUW01000002 SPP77549.1 KZ288400 PBC26288.1 CH902618 EDV39021.1 CH954178 EDV50977.1 KQ982877 KYQ49700.1 GFTR01000772 JAW15654.1 GL888147 EGI66559.1 KQ981724 KYN37240.1 CM000159 EDW93469.1 ACPB03010999 GECZ01000211 JAS69558.1 AE014296 AY084166 KX531355 AAF47950.2 AAL89904.1 ANY27165.1 CM000363 CM002912 EDX09275.1 KMY97691.1 CH480817 EDW50541.1 KQ981063 KYN09959.1 JRES01001653 KNC21127.1 GFDF01010780 JAV03304.1 KQ435912 KOX68895.1 CP012525 ALC43065.1 GFDF01010927 JAV03157.1 GFDF01010926 JAV03158.1 CCAG010011675
Proteomes
UP000053240
UP000053268
UP000283053
UP000008820
UP000002320
UP000069940
+ More
UP000249989 UP000075886 UP000075900 UP000007062 UP000075920 UP000075903 UP000000673 UP000076502 UP000053825 UP000075880 UP000092462 UP000279307 UP000008237 UP000095301 UP000002358 UP000215335 UP000027135 UP000183832 UP000007798 UP000005203 UP000030765 UP000075885 UP000005205 UP000095300 UP000192223 UP000036403 UP000076408 UP000075840 UP000007266 UP000069272 UP000076407 UP000075884 UP000075881 UP000001070 UP000000311 UP000008792 UP000009192 UP000078540 UP000001819 UP000008744 UP000268350 UP000242457 UP000007801 UP000008711 UP000075809 UP000007755 UP000192221 UP000078541 UP000002282 UP000015103 UP000000803 UP000000304 UP000001292 UP000078492 UP000037069 UP000053105 UP000092553 UP000079169 UP000092445 UP000092444
UP000249989 UP000075886 UP000075900 UP000007062 UP000075920 UP000075903 UP000000673 UP000076502 UP000053825 UP000075880 UP000092462 UP000279307 UP000008237 UP000095301 UP000002358 UP000215335 UP000027135 UP000183832 UP000007798 UP000005203 UP000030765 UP000075885 UP000005205 UP000095300 UP000192223 UP000036403 UP000076408 UP000075840 UP000007266 UP000069272 UP000076407 UP000075884 UP000075881 UP000001070 UP000000311 UP000008792 UP000009192 UP000078540 UP000001819 UP000008744 UP000268350 UP000242457 UP000007801 UP000008711 UP000075809 UP000007755 UP000192221 UP000078541 UP000002282 UP000015103 UP000000803 UP000000304 UP000001292 UP000078492 UP000037069 UP000053105 UP000092553 UP000079169 UP000092445 UP000092444
PRIDE
Pfam
Interpro
IPR035699
AAA_6
+ More
IPR042228 Dynein_2_C
IPR041228 Dynein_C
IPR041658 AAA_lid_11
IPR003593 AAA+_ATPase
IPR041466 Dynein_AAA5_ext
IPR013602 Dynein_heavy_dom-2
IPR004273 Dynein_heavy_D6_P-loop
IPR026983 DHC_fam
IPR042219 AAA_lid_11_sf
IPR024743 Dynein_HC_stalk
IPR024317 Dynein_heavy_chain_D4_dom
IPR027417 P-loop_NTPase
IPR042222 Dynein_2_N
IPR041589 DNAH3_AAA_lid_1
IPR035706 AAA_9
IPR037294 ABC_BtuC-like
IPR004019 YLP_motif
IPR004011 Gyr_motif
IPR042228 Dynein_2_C
IPR041228 Dynein_C
IPR041658 AAA_lid_11
IPR003593 AAA+_ATPase
IPR041466 Dynein_AAA5_ext
IPR013602 Dynein_heavy_dom-2
IPR004273 Dynein_heavy_D6_P-loop
IPR026983 DHC_fam
IPR042219 AAA_lid_11_sf
IPR024743 Dynein_HC_stalk
IPR024317 Dynein_heavy_chain_D4_dom
IPR027417 P-loop_NTPase
IPR042222 Dynein_2_N
IPR041589 DNAH3_AAA_lid_1
IPR035706 AAA_9
IPR037294 ABC_BtuC-like
IPR004019 YLP_motif
IPR004011 Gyr_motif
Gene 3D
ProteinModelPortal
A0A194QTZ8
A0A194PJ13
A0A3S2LR59
Q16ZV7
B0WXN7
A0A023ECJ5
+ More
A0A1Q3FL49 A0A1Q3FKU0 A0A182QWW1 A0A182R6C8 Q7PYM4 A0A182VZ14 A0A182V5Y7 W5JJ86 T1EAP0 A0A154PSP8 A0A0L7R2Y7 A0A182JK68 U5ED05 A0A336M033 A0A1B0D512 A0A1Y1LH00 A0A3L8DD20 E2C4G1 A0A1I8M343 K7JGT6 A0A232ERU0 A0A067QXH5 A0A1J1IG39 B4MMW4 A0A088A5M0 A0A1L8EB14 A0A084VHD4 A0A182PKK7 A0A1L8EG28 A0A158NFG7 A0A1I8PJ61 A0A1W4W405 A0A310SLD9 F5HM97 A0A0J7KJF3 A0A182XVX1 E9ITG7 A0A182I057 A0A139WLL4 A0A182FKT1 A0A182XBB4 A0A182NCF2 A0A182JWP4 B4IZ57 E2A6R5 B4MG12 B4KXC6 A0A195BDK4 Q29E95 B4HD20 A0A3B0JV09 A0A2A3E3M4 B3M9L5 B3NG76 A0A151WPE6 A0A224XWP0 F4WGU0 A0A1W4VLR6 A0A151JVB0 B4PIC7 T1HX73 A0A1B6H4G4 Q8SXS4 B4QR38 B4HUC0 A0A195DC25 A0A0L0BM80 A0A1L8DAF8 A0A0M8ZQ93 A0A0M4EFL2 A0A1S3DFQ1 A0A1A9ZUE8 A0A1L8D9X0 A0A1L8D9L1 A0A1B0FC04
A0A1Q3FL49 A0A1Q3FKU0 A0A182QWW1 A0A182R6C8 Q7PYM4 A0A182VZ14 A0A182V5Y7 W5JJ86 T1EAP0 A0A154PSP8 A0A0L7R2Y7 A0A182JK68 U5ED05 A0A336M033 A0A1B0D512 A0A1Y1LH00 A0A3L8DD20 E2C4G1 A0A1I8M343 K7JGT6 A0A232ERU0 A0A067QXH5 A0A1J1IG39 B4MMW4 A0A088A5M0 A0A1L8EB14 A0A084VHD4 A0A182PKK7 A0A1L8EG28 A0A158NFG7 A0A1I8PJ61 A0A1W4W405 A0A310SLD9 F5HM97 A0A0J7KJF3 A0A182XVX1 E9ITG7 A0A182I057 A0A139WLL4 A0A182FKT1 A0A182XBB4 A0A182NCF2 A0A182JWP4 B4IZ57 E2A6R5 B4MG12 B4KXC6 A0A195BDK4 Q29E95 B4HD20 A0A3B0JV09 A0A2A3E3M4 B3M9L5 B3NG76 A0A151WPE6 A0A224XWP0 F4WGU0 A0A1W4VLR6 A0A151JVB0 B4PIC7 T1HX73 A0A1B6H4G4 Q8SXS4 B4QR38 B4HUC0 A0A195DC25 A0A0L0BM80 A0A1L8DAF8 A0A0M8ZQ93 A0A0M4EFL2 A0A1S3DFQ1 A0A1A9ZUE8 A0A1L8D9X0 A0A1L8D9L1 A0A1B0FC04
Ontologies
PANTHER
Topology
SignalP
Position: 1 - 27,
Likelihood: 0.593977
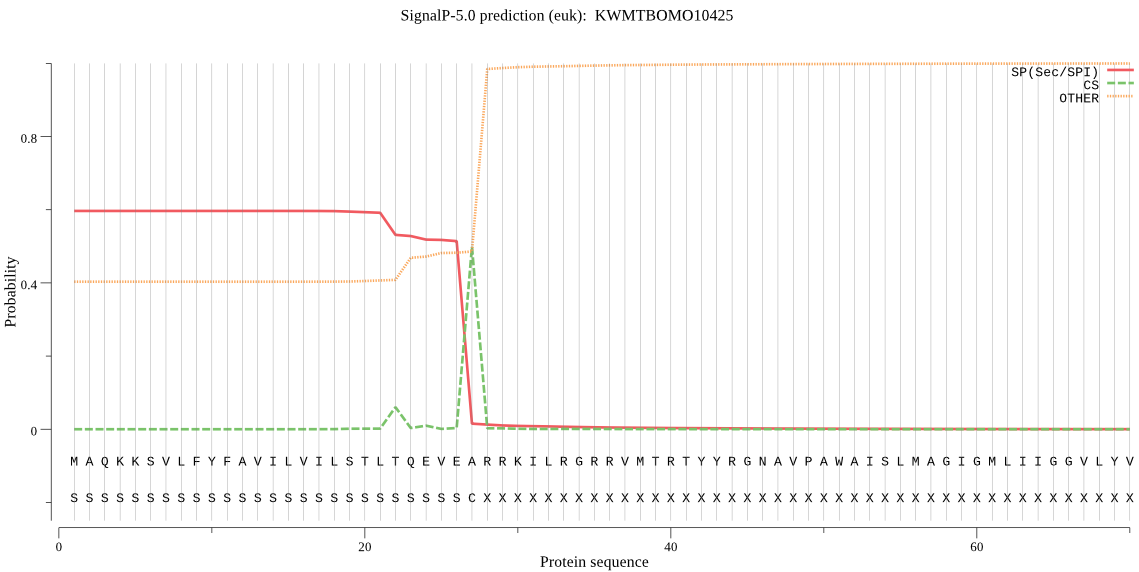
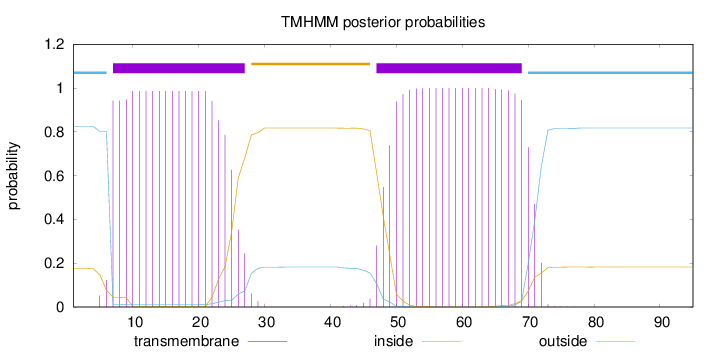
Length:
95
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
41.59774
Exp number, first 60 AAs:
31.27665
Total prob of N-in:
0.82484
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 27
outside
28 - 46
TMhelix
47 - 69
inside
70 - 95
Population Genetic Test Statistics
Pi
271.130182
Theta
210.748351
Tajima's D
0.866871
CLR
0.020798
CSRT
0.621568921553922
Interpretation
Uncertain
