Gene
KWMTBOMO10421
Pre Gene Modal
BGIBMGA007081
Annotation
PREDICTED:_DNA_topoisomerase_3-alpha_[Bombyx_mori]
Full name
DNA topoisomerase
+ More
DNA topoisomerase 3-alpha
DNA topoisomerase 3-alpha
Alternative Name
DNA topoisomerase III alpha
Location in the cell
Nuclear Reliability : 2.612
Sequence
CDS
ATGAAATATTTAAATGTAGCAGAGAAAAATGATGCAGCTAAAAATATTGCATCACATCTATCCAAAGGAACATCAACTGGGCGTAAAGGACTATCAAAGTACAACAAAATATATCAATTTAAAGCCAACGTAATGGGGCAAGATTGTGAAATGGTGATGACCTCAGTATCAGGACATCTCTTAGCCCTAGAATTTAGTGGAACATACAGAACCTGGCAAACCTGTAGCCCACTATCACTGTTTGATGCACCTGTTTTCAAATACTGCCCAGAGAATTACCAAAAAATTAAGATGACCTTAGAGCGAGAAGTTCGAAGCTGTCAAGGCCTCATAATATGGACGGATTGTGACAGAGAAGGAGAGAACATAGGTTTTGAGATTATTGATGTTTGTAAAGCTGCCAGGACAAATTTGAAAATATATAGAGCAAAATTTTCAGAGATCACATTAGTTTCAGTAAATAGAGCTTTACAAAACTTAGAAGAACCAAACAAGAATACTAGTAATGCAGTAGATGTCAGACAAGAACTTGATTTAAGGATAGGTGCGGCATTCACAAGGTTCCAAACATTGAGACTCCAAAAGGTATTCCCAAGTAAACTAACAGACACCATAGTGAGTTATGGTTCTTGTCAATTTCCCACTCTTGGATTTGTGGTGGAACGCTATAGAGCCATAGAAAACTTCATTGTCGAAACGTTTTGGAAATTAAAGGTAAATCATACTATAGAGAACCTCAGTGTAGAGTTCGCATGGGAACGTTTTCGGTTGTTTGACGAATATGCTTGTAAAATATTTCATGATATTTGCACTGAGAATCCTTTGGCGAGAGTGCTTGAAGTGAAAACTAAACCTAAGTCTAAATGGCGGCCGTTGCCTTTGGAAACTGTGGAACTAGAAAAGCAAGCGTCACGTAAATTAAAAATACACGCGAAAGAGACAATGCAACTCGCCGAGAAACTCTACACCCAAGGCTTCATCAGCTACCCTCGTACAGAGACAAATGAGTTTCCGAAGGAGATGAACTTAGCCCAGTTAGTTGGTCAGCAGACCGGTGATCCGAACTGGGGTACATTCGCGCAGAAGATATTAGATAACGGCGGGCCAACTCCGAGGCAAGGAAAGAAAAGCGACAAAGCGCATCCGCCTATACATCCAACAAAATATACGAACAATTTATCGGGCAACGAGAAACGCCTTTACGAGTTCATAGTGCGATCGTTCCTTGCGTGCTGTTCCAAGGACGCTCAGGGTCAGGAGACGATCGTTAACATAGATATAGGCAGAGAGAAGTTTTCCGCCAACGGACTCGTGATCACCGAATTAAACTACCTAGAGGTATACATCTACGAGAAGTGGTCGTCTAAAGAAATACACGTCTATCAGGAAGGTCAAATATTCAGTCCAACCTCAATAGACATCGTATCCGGTACTACTTCACCTCCTAGCTTACTAACAGAAGCCGACCTCATTAGTCTCATGGAAAAACATGGCATCGGGACAGACGCAACGCACGCTGAGCACATTGAGAAGATAAAAAGCCGTTCGTATGTAGCGTTGGCAGATGAAAAATATTTCGTTCCCGGTGTCTTAGGCATGGGACTGGTGGAGGGCTACGATGCTATGGGCCTCGAAATGTCGAAGCCTCATCTCCGTGCTCAGTTAGAGTCTGATCTCAAAGAAATTATCGAAGGAAGGAAACAGCCTGAAGCGGTTTTATCTGAACAGATAGCAAAATATAAAGAGGTGTACATTAAAGCATCGGCGGAGGCTGAGAAGATAGACAGAGCGCTCGCGGAGAGACTGAACGAGCAGCCGGCGGCGCCGCCTGCGAGCGGGGATACCTTACCGGTCGACATGCCCGCGGCCTTAAAATGTCCGAAATGTGGCTCGGACATGATCGTAAGACAGAAAAAGAATAATCCGACCGAATATTATATCGGTTGCATGTCATTTCCTAACTGCAAAAATGTAGTTTGGCTACCTAATATAGTTAAGTCATTGCAAGTTTTACCAGACACTTGCACAGAGTGTGGACCGGACTACAAGATGATGAAGTTTGAATGGCGAAACAATTCCATAAGTCACATATACGCACCGCCGTACACGGGCTGCATCGGCGGGTGCGATAGAACGTTTCTAGAATATCTCAATATAAACATCAACAGTGTCATAAAAACATCGGGCTCCTCCGCACAAGTCACCTCTTCGAACACCAGTTCGAGAACAGATATATCAGTACCCATACAAAACCATATTATACCGAGTAATGCACTTCGAAATCCCATAGAAATTGGTTCAAATAGAGGTAGAGATATACCGGCGCAGAGGAACAATGCTATGAACACGACAAACATTAGGATGAACAGCGCCAGTAATGCCGCTAACTTAGATGACAATCACGTGGTCTGCAGATGCTCAAAACCGGCGATCTTATTAACGTCTAAGAAACAAAATCAGAATTTTGGCCAAAAATTTTACAAATGTCCAGTCGGGAAAGAGAACGGTGGATGTGATTTCTTCTTGTGGGCTCCAGAATCCGTCAATAATGCTCAGTCTGCATTCGAGACTCCGCCGTCGTCGTCTGAGCAAACGACTTTTAATACAACTGGTGTTGCCAGACGGGATGCAAATCGTAATAGAAATACTGGGTCTGGCAACACTGATGACGTCAAGTGTGACTGTCGTTTGCCTTGTAAAAAACTAACGGTACACAAAGAGGGCCCAAATAAAGGACGACAGTTCTATGGGTGCGCGAAAGACTTGAGCTCGAAATGCAACTTCTTTAAATGGGCCGATGAAAGCGGCTCCAGACCAGGTCCTGCTCGGCCTGCCCCTGTGAGCTCCCGCGGCGGAGGCCGAGCGTCCCGCCGGGGCGCGGTGCGGCATTGCAGCTTCTGTGGGAGCACCGAACACGACTTTCGAACCTGCTCTATAAGGAGACCCTCCACAGACTAA
Protein
MKYLNVAEKNDAAKNIASHLSKGTSTGRKGLSKYNKIYQFKANVMGQDCEMVMTSVSGHLLALEFSGTYRTWQTCSPLSLFDAPVFKYCPENYQKIKMTLEREVRSCQGLIIWTDCDREGENIGFEIIDVCKAARTNLKIYRAKFSEITLVSVNRALQNLEEPNKNTSNAVDVRQELDLRIGAAFTRFQTLRLQKVFPSKLTDTIVSYGSCQFPTLGFVVERYRAIENFIVETFWKLKVNHTIENLSVEFAWERFRLFDEYACKIFHDICTENPLARVLEVKTKPKSKWRPLPLETVELEKQASRKLKIHAKETMQLAEKLYTQGFISYPRTETNEFPKEMNLAQLVGQQTGDPNWGTFAQKILDNGGPTPRQGKKSDKAHPPIHPTKYTNNLSGNEKRLYEFIVRSFLACCSKDAQGQETIVNIDIGREKFSANGLVITELNYLEVYIYEKWSSKEIHVYQEGQIFSPTSIDIVSGTTSPPSLLTEADLISLMEKHGIGTDATHAEHIEKIKSRSYVALADEKYFVPGVLGMGLVEGYDAMGLEMSKPHLRAQLESDLKEIIEGRKQPEAVLSEQIAKYKEVYIKASAEAEKIDRALAERLNEQPAAPPASGDTLPVDMPAALKCPKCGSDMIVRQKKNNPTEYYIGCMSFPNCKNVVWLPNIVKSLQVLPDTCTECGPDYKMMKFEWRNNSISHIYAPPYTGCIGGCDRTFLEYLNININSVIKTSGSSAQVTSSNTSSRTDISVPIQNHIIPSNALRNPIEIGSNRGRDIPAQRNNAMNTTNIRMNSASNAANLDDNHVVCRCSKPAILLTSKKQNQNFGQKFYKCPVGKENGGCDFFLWAPESVNNAQSAFETPPSSSEQTTFNTTGVARRDANRNRNTGSGNTDDVKCDCRLPCKKLTVHKEGPNKGRQFYGCAKDLSSKCNFFKWADESGSRPGPARPAPVSSRGGGRASRRGAVRHCSFCGSTEHDFRTCSIRRPSTD
Summary
Description
Introduces a single-strand break via transesterification at a target site in duplex DNA. Releases the supercoiling and torsional tension of DNA introduced during the DNA replication and transcription by transiently cleaving and rejoining one strand of the DNA duplex. The scissile phosphodiester is attacked by the catalytic tyrosine of the enzyme, resulting in the formation of a DNA-(5'-phosphotyrosyl)-enzyme intermediate and the expulsion of a 3'-OH DNA strand.
Releases the supercoiling and torsional tension of DNA introduced during the DNA replication and transcription by transiently cleaving and rejoining one strand of the DNA duplex. Introduces a single-strand break via transesterification at a target site in duplex DNA. The scissile phosphodiester is attacked by the catalytic tyrosine of the enzyme, resulting in the formation of a DNA-(5'-phosphotyrosyl)-enzyme intermediate and the expulsion of a 3'-OH DNA strand. The free DNA strand then undergoes passage around the unbroken strand thus removing DNA supercoils. Finally, in the religation step, the DNA 3'-OH attacks the covalent intermediate to expel the active-site tyrosine and restore the DNA phosphodiester backbone. As an essential component of the RMI complex it is involved in chromosome separation and the processing of homologous recombination intermediates to limit DNA crossover formation in cells. Has DNA decatenation activity. It is required for mtDNA decatenation and segregation after completion of replication, in a process that does not require BLM, RMI1 and RMI2.
Releases the supercoiling and torsional tension of DNA introduced during the DNA replication and transcription by transiently cleaving and rejoining one strand of the DNA duplex. Introduces a single-strand break via transesterification at a target site in duplex DNA. The scissile phosphodiester is attacked by the catalytic tyrosine of the enzyme, resulting in the formation of a DNA-(5'-phosphotyrosyl)-enzyme intermediate and the expulsion of a 3'-OH DNA strand. The free DNA strand then undergoes passage around the unbroken strand thus removing DNA supercoils. Finally, in the religation step, the DNA 3'-OH attacks the covalent intermediate to expel the active-site tyrosine and restore the DNA phosphodiester backbone. As an essential component of the RMI complex it is involved in chromosome separation and the processing of homologous recombination intermediates to limit DNA crossover formation in cells. Has DNA decatenation activity. It is required for mtDNA decatenation and segregation after completion of replication, in a process that does not require BLM, RMI1 and RMI2.
Catalytic Activity
ATP-independent breakage of single-stranded DNA, followed by passage and rejoining.
Cofactor
Mg(2+)
Subunit
Binds ssDNA. Interacts (via N-terminal region) with BLM; the interaction is direct. Directly interacts with RMI1. Component of the RMI complex, containing at least TOP3A, RMI1 and RMI2. The RMI complex interacts with BLM.
Similarity
Belongs to the type IA topoisomerase family.
Keywords
Complete proteome
DNA-binding
Isomerase
Magnesium
Metal-binding
Mitochondrion
Reference proteome
Repeat
Topoisomerase
Zinc
Zinc-finger
Feature
chain DNA topoisomerase 3-alpha
Uniprot
H9JC36
A0A3S2NCN1
A0A2A4IYV0
A0A194PJY1
A0A194QTZ3
A0A2H1VAY8
+ More
A0A2J7QL57 A0A2J7QL50 A0A0L7RD08 D6WYH2 A0A0J7K841 A0A091UUX7 A0A091EPK2 U3KH13 A0A087V368 A0A091IBC6 A0A091PE50 A0A195F5T7 E2A6W4 A0A091UJS7 F1P1F3 H3AKB8 A0A091VY07 A0A0A0AQJ5 Q4W5X0 A0A091U1D3 A0A094NN34 A0A1V4KSI4 A0A091S7V4 W5KS91 A0A093F4L3 A0A093LKR5 A0A091KTW7 A0A1S3F5Y4 A0A3P9KGI1 A0A226PP95 X1WCF3 A0A091NCS3 A0A0M9A9F6 A0A087ZSA7 A0A093H5G5 H9GF12 A0A091MVC1 R0JZT4 A0A1B6E1A8 A0A226NNR0 A0A094LFW4 A0A3P9KGC2 A0A3N0YUR3 H2MFK0 A0A3B4CZI1 Q5NCT2 O70157 A0A3B4CZJ8 Q6GPC0 A0A091VA01 A0A2I0LTH8 A0A3B3IA27 A0A091G737 K7GGN4 A0A3B3HFK0 G1N063 A0A093R579 A0A093C8L8 Q3V3P1 A0A3Q3L822 I3JP28 A0A2Y9HEV5 H0Z9W6 A0A1L8EYN9 K7GGM4 A0A087RG32 A0A091ISF3 A0A2D0PXW3 A0A0Q3XAU4 I3JP29 A0A3B3BFF3 A0A3P9IEV5 A0A3P8PWM6 A0A3P8PWH6 H0VEI8 A0A3Q2VSU4 F7A1S4 K9INH6 A0A3B4HD57 A0A093FX84 M3XLT6 A0A3P9B4D0 A0A3Q7NG50 A0A093QKD6 A0A287D6R7 N6UAY1 I3MP50 A0A091DBP4 G1NSV2 A0A2I3GKX0 A0A2U9CPT7 S7PHJ6 A0A093D0I4 A0A2K6KS94
A0A2J7QL57 A0A2J7QL50 A0A0L7RD08 D6WYH2 A0A0J7K841 A0A091UUX7 A0A091EPK2 U3KH13 A0A087V368 A0A091IBC6 A0A091PE50 A0A195F5T7 E2A6W4 A0A091UJS7 F1P1F3 H3AKB8 A0A091VY07 A0A0A0AQJ5 Q4W5X0 A0A091U1D3 A0A094NN34 A0A1V4KSI4 A0A091S7V4 W5KS91 A0A093F4L3 A0A093LKR5 A0A091KTW7 A0A1S3F5Y4 A0A3P9KGI1 A0A226PP95 X1WCF3 A0A091NCS3 A0A0M9A9F6 A0A087ZSA7 A0A093H5G5 H9GF12 A0A091MVC1 R0JZT4 A0A1B6E1A8 A0A226NNR0 A0A094LFW4 A0A3P9KGC2 A0A3N0YUR3 H2MFK0 A0A3B4CZI1 Q5NCT2 O70157 A0A3B4CZJ8 Q6GPC0 A0A091VA01 A0A2I0LTH8 A0A3B3IA27 A0A091G737 K7GGN4 A0A3B3HFK0 G1N063 A0A093R579 A0A093C8L8 Q3V3P1 A0A3Q3L822 I3JP28 A0A2Y9HEV5 H0Z9W6 A0A1L8EYN9 K7GGM4 A0A087RG32 A0A091ISF3 A0A2D0PXW3 A0A0Q3XAU4 I3JP29 A0A3B3BFF3 A0A3P9IEV5 A0A3P8PWM6 A0A3P8PWH6 H0VEI8 A0A3Q2VSU4 F7A1S4 K9INH6 A0A3B4HD57 A0A093FX84 M3XLT6 A0A3P9B4D0 A0A3Q7NG50 A0A093QKD6 A0A287D6R7 N6UAY1 I3MP50 A0A091DBP4 G1NSV2 A0A2I3GKX0 A0A2U9CPT7 S7PHJ6 A0A093D0I4 A0A2K6KS94
EC Number
5.6.2.2
Pubmed
EMBL
BABH01014822
BABH01014823
BABH01014824
BABH01014825
BABH01014826
RSAL01000097
+ More
RVE47675.1 NWSH01004560 PCG64931.1 KQ459603 KPI93034.1 KQ461137 KPJ08784.1 ODYU01001340 SOQ37384.1 NEVH01013252 PNF29310.1 PNF29311.1 KQ414615 KOC68743.1 KQ971362 EFA08981.1 LBMM01012048 KMQ86449.1 KK465315 KFQ81468.1 KK718885 KFO58976.1 AGTO01020911 KL478123 KFO07060.1 KL218418 KFP04783.1 KK656909 KFQ05538.1 KQ981805 KYN35542.1 GL437203 EFN70830.1 KK434863 KFQ89960.1 AADN05000376 AFYH01084656 AFYH01084657 AFYH01084658 KK734455 KFR08069.1 KL872578 KGL96157.1 AB215104 BAD99110.1 KK494841 KFQ64986.1 KL282336 KFZ67856.1 LSYS01001700 OPJ87403.1 KK945168 KFQ53766.1 KK621420 KFV51470.1 KK606573 KFW09486.1 KK754018 KFP42710.1 AWGT02000031 OXB81916.1 BX649440 BX649547 KK845599 KFP87266.1 KQ435714 KOX79196.1 KL205998 KFV77823.1 AAWZ02031062 KK518124 KFP65450.1 KB742883 EOB03181.1 GEDC01005607 JAS31691.1 MCFN01000008 OXB68779.1 KL357022 KFZ62874.1 RJVU01026577 ROL49438.1 BC099689 AAH99689.1 AB006074 BC073221 AAH73221.1 KL410807 KFQ99903.1 AKCR02000102 PKK20723.1 KL447789 KFO77141.1 AGCU01109724 AGCU01109725 AGCU01109726 AGCU01109727 KL439394 KFW93790.1 KL235497 KFV08617.1 AL596215 AK037640 BAE20515.1 AERX01009134 AERX01009135 ABQF01123673 CM004482 OCT64461.1 KL226347 KFM12436.1 KK500930 KFP11629.1 LMAW01000001 KQL61419.1 AAKN02045141 AAMC01050874 GABZ01004040 JAA49485.1 KL214742 KFV61393.1 AEYP01089335 AEYP01089336 AEYP01089337 KL672051 KFW84487.1 AGTP01101833 AGTP01101834 AGTP01101835 APGK01036017 APGK01036018 KB740932 ENN77811.1 KN122870 KFO27670.1 AAPE02048301 AAPE02048302 AAPE02048303 ADFV01121230 ADFV01121231 ADFV01121232 ADFV01121233 ADFV01121234 ADFV01121235 ADFV01121236 ADFV01121237 CP026261 AWP18598.1 KE162945 EPQ10148.1 KL474997 KFV20288.1
RVE47675.1 NWSH01004560 PCG64931.1 KQ459603 KPI93034.1 KQ461137 KPJ08784.1 ODYU01001340 SOQ37384.1 NEVH01013252 PNF29310.1 PNF29311.1 KQ414615 KOC68743.1 KQ971362 EFA08981.1 LBMM01012048 KMQ86449.1 KK465315 KFQ81468.1 KK718885 KFO58976.1 AGTO01020911 KL478123 KFO07060.1 KL218418 KFP04783.1 KK656909 KFQ05538.1 KQ981805 KYN35542.1 GL437203 EFN70830.1 KK434863 KFQ89960.1 AADN05000376 AFYH01084656 AFYH01084657 AFYH01084658 KK734455 KFR08069.1 KL872578 KGL96157.1 AB215104 BAD99110.1 KK494841 KFQ64986.1 KL282336 KFZ67856.1 LSYS01001700 OPJ87403.1 KK945168 KFQ53766.1 KK621420 KFV51470.1 KK606573 KFW09486.1 KK754018 KFP42710.1 AWGT02000031 OXB81916.1 BX649440 BX649547 KK845599 KFP87266.1 KQ435714 KOX79196.1 KL205998 KFV77823.1 AAWZ02031062 KK518124 KFP65450.1 KB742883 EOB03181.1 GEDC01005607 JAS31691.1 MCFN01000008 OXB68779.1 KL357022 KFZ62874.1 RJVU01026577 ROL49438.1 BC099689 AAH99689.1 AB006074 BC073221 AAH73221.1 KL410807 KFQ99903.1 AKCR02000102 PKK20723.1 KL447789 KFO77141.1 AGCU01109724 AGCU01109725 AGCU01109726 AGCU01109727 KL439394 KFW93790.1 KL235497 KFV08617.1 AL596215 AK037640 BAE20515.1 AERX01009134 AERX01009135 ABQF01123673 CM004482 OCT64461.1 KL226347 KFM12436.1 KK500930 KFP11629.1 LMAW01000001 KQL61419.1 AAKN02045141 AAMC01050874 GABZ01004040 JAA49485.1 KL214742 KFV61393.1 AEYP01089335 AEYP01089336 AEYP01089337 KL672051 KFW84487.1 AGTP01101833 AGTP01101834 AGTP01101835 APGK01036017 APGK01036018 KB740932 ENN77811.1 KN122870 KFO27670.1 AAPE02048301 AAPE02048302 AAPE02048303 ADFV01121230 ADFV01121231 ADFV01121232 ADFV01121233 ADFV01121234 ADFV01121235 ADFV01121236 ADFV01121237 CP026261 AWP18598.1 KE162945 EPQ10148.1 KL474997 KFV20288.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000053240
UP000235965
+ More
UP000053825 UP000007266 UP000036403 UP000052976 UP000016665 UP000054308 UP000078541 UP000000311 UP000000539 UP000008672 UP000053605 UP000053858 UP000190648 UP000018467 UP000081671 UP000265180 UP000198419 UP000000437 UP000053105 UP000005203 UP000053584 UP000001646 UP000198323 UP000001038 UP000261440 UP000000589 UP000053283 UP000053872 UP000053760 UP000007267 UP000001645 UP000261640 UP000005207 UP000248481 UP000007754 UP000186698 UP000053286 UP000053119 UP000221080 UP000051836 UP000261560 UP000265200 UP000265100 UP000005447 UP000264840 UP000008143 UP000261460 UP000053875 UP000000715 UP000265160 UP000286641 UP000053258 UP000005215 UP000019118 UP000028990 UP000001074 UP000001073 UP000246464 UP000233180
UP000053825 UP000007266 UP000036403 UP000052976 UP000016665 UP000054308 UP000078541 UP000000311 UP000000539 UP000008672 UP000053605 UP000053858 UP000190648 UP000018467 UP000081671 UP000265180 UP000198419 UP000000437 UP000053105 UP000005203 UP000053584 UP000001646 UP000198323 UP000001038 UP000261440 UP000000589 UP000053283 UP000053872 UP000053760 UP000007267 UP000001645 UP000261640 UP000005207 UP000248481 UP000007754 UP000186698 UP000053286 UP000053119 UP000221080 UP000051836 UP000261560 UP000265200 UP000265100 UP000005447 UP000264840 UP000008143 UP000261460 UP000053875 UP000000715 UP000265160 UP000286641 UP000053258 UP000005215 UP000019118 UP000028990 UP000001074 UP000001073 UP000246464 UP000233180
Interpro
IPR003601
Topo_IA_2
+ More
IPR006171 TOPRIM_domain
IPR013497 Topo_IA_cen
IPR013826 Topo_IA_cen_sub3
IPR023405 Topo_IA_core_domain
IPR034144 TOPRIM_TopoIII
IPR023406 Topo_IA_AS
IPR013498 Topo_IA_Znf
IPR003602 Topo_IA_DNA-bd_dom
IPR013824 Topo_IA_cen_sub1
IPR013825 Topo_IA_cen_sub2
IPR010666 Znf_GRF
IPR000380 Topo_IA
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR006171 TOPRIM_domain
IPR013497 Topo_IA_cen
IPR013826 Topo_IA_cen_sub3
IPR023405 Topo_IA_core_domain
IPR034144 TOPRIM_TopoIII
IPR023406 Topo_IA_AS
IPR013498 Topo_IA_Znf
IPR003602 Topo_IA_DNA-bd_dom
IPR013824 Topo_IA_cen_sub1
IPR013825 Topo_IA_cen_sub2
IPR010666 Znf_GRF
IPR000380 Topo_IA
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
Gene 3D
ProteinModelPortal
H9JC36
A0A3S2NCN1
A0A2A4IYV0
A0A194PJY1
A0A194QTZ3
A0A2H1VAY8
+ More
A0A2J7QL57 A0A2J7QL50 A0A0L7RD08 D6WYH2 A0A0J7K841 A0A091UUX7 A0A091EPK2 U3KH13 A0A087V368 A0A091IBC6 A0A091PE50 A0A195F5T7 E2A6W4 A0A091UJS7 F1P1F3 H3AKB8 A0A091VY07 A0A0A0AQJ5 Q4W5X0 A0A091U1D3 A0A094NN34 A0A1V4KSI4 A0A091S7V4 W5KS91 A0A093F4L3 A0A093LKR5 A0A091KTW7 A0A1S3F5Y4 A0A3P9KGI1 A0A226PP95 X1WCF3 A0A091NCS3 A0A0M9A9F6 A0A087ZSA7 A0A093H5G5 H9GF12 A0A091MVC1 R0JZT4 A0A1B6E1A8 A0A226NNR0 A0A094LFW4 A0A3P9KGC2 A0A3N0YUR3 H2MFK0 A0A3B4CZI1 Q5NCT2 O70157 A0A3B4CZJ8 Q6GPC0 A0A091VA01 A0A2I0LTH8 A0A3B3IA27 A0A091G737 K7GGN4 A0A3B3HFK0 G1N063 A0A093R579 A0A093C8L8 Q3V3P1 A0A3Q3L822 I3JP28 A0A2Y9HEV5 H0Z9W6 A0A1L8EYN9 K7GGM4 A0A087RG32 A0A091ISF3 A0A2D0PXW3 A0A0Q3XAU4 I3JP29 A0A3B3BFF3 A0A3P9IEV5 A0A3P8PWM6 A0A3P8PWH6 H0VEI8 A0A3Q2VSU4 F7A1S4 K9INH6 A0A3B4HD57 A0A093FX84 M3XLT6 A0A3P9B4D0 A0A3Q7NG50 A0A093QKD6 A0A287D6R7 N6UAY1 I3MP50 A0A091DBP4 G1NSV2 A0A2I3GKX0 A0A2U9CPT7 S7PHJ6 A0A093D0I4 A0A2K6KS94
A0A2J7QL57 A0A2J7QL50 A0A0L7RD08 D6WYH2 A0A0J7K841 A0A091UUX7 A0A091EPK2 U3KH13 A0A087V368 A0A091IBC6 A0A091PE50 A0A195F5T7 E2A6W4 A0A091UJS7 F1P1F3 H3AKB8 A0A091VY07 A0A0A0AQJ5 Q4W5X0 A0A091U1D3 A0A094NN34 A0A1V4KSI4 A0A091S7V4 W5KS91 A0A093F4L3 A0A093LKR5 A0A091KTW7 A0A1S3F5Y4 A0A3P9KGI1 A0A226PP95 X1WCF3 A0A091NCS3 A0A0M9A9F6 A0A087ZSA7 A0A093H5G5 H9GF12 A0A091MVC1 R0JZT4 A0A1B6E1A8 A0A226NNR0 A0A094LFW4 A0A3P9KGC2 A0A3N0YUR3 H2MFK0 A0A3B4CZI1 Q5NCT2 O70157 A0A3B4CZJ8 Q6GPC0 A0A091VA01 A0A2I0LTH8 A0A3B3IA27 A0A091G737 K7GGN4 A0A3B3HFK0 G1N063 A0A093R579 A0A093C8L8 Q3V3P1 A0A3Q3L822 I3JP28 A0A2Y9HEV5 H0Z9W6 A0A1L8EYN9 K7GGM4 A0A087RG32 A0A091ISF3 A0A2D0PXW3 A0A0Q3XAU4 I3JP29 A0A3B3BFF3 A0A3P9IEV5 A0A3P8PWM6 A0A3P8PWH6 H0VEI8 A0A3Q2VSU4 F7A1S4 K9INH6 A0A3B4HD57 A0A093FX84 M3XLT6 A0A3P9B4D0 A0A3Q7NG50 A0A093QKD6 A0A287D6R7 N6UAY1 I3MP50 A0A091DBP4 G1NSV2 A0A2I3GKX0 A0A2U9CPT7 S7PHJ6 A0A093D0I4 A0A2K6KS94
PDB
4CGY
E-value=0,
Score=2310
Ontologies
PATHWAY
GO
PANTHER
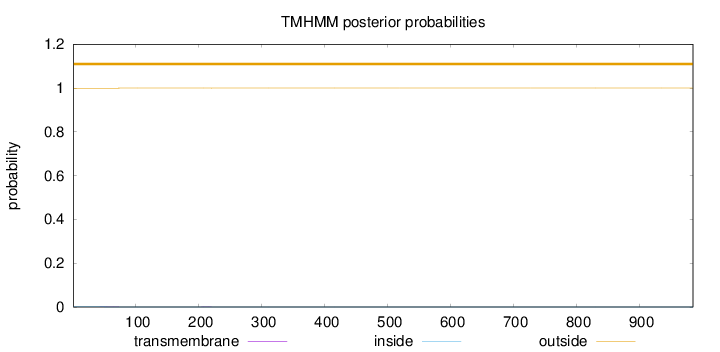
Topology
Subcellular location
Mitochondrion matrix
Length:
985
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02387
Exp number, first 60 AAs:
0.01013
Total prob of N-in:
0.00117
outside
1 - 985
Population Genetic Test Statistics
Pi
227.522845
Theta
195.739135
Tajima's D
0.800512
CLR
0.073052
CSRT
0.607619619019049
Interpretation
Uncertain
