Gene
KWMTBOMO10420
Pre Gene Modal
BGIBMGA007021
Annotation
hypothetical_protein_KGM_19075_[Danaus_plexippus]
Full name
Serine hydroxymethyltransferase
Location in the cell
Nuclear Reliability : 2.652
Sequence
CDS
ATGACGATGTTTCCTGCTTCTGGATCTGCTGCGGTGCCGCAGCAAACAATAAATGGGCAAGTAGTGCAAAACGGCAGCACGACTTTGCCTCGCTCCCATCACCAGAGAACAGGGCGACCACCGGCAGCTCCTAAATCATATGATTACCTTTTAAAAGTGCTACTAGTTGGTGATTCTGATGTTGGAAAACAAGAGATCCTTCAAGATTTAGAGGATGGCTCTGCAGATTCACCCTTTTGCAGCGGTAGTGGATACAAGACCACGACAATACTTTTGGACGGAAAGAGAGTTAAGCTTCAGCTATGGGATACCTCAGGACAGGGAAGGTTTTGTACTATAATTCGGTCATACTCACGTGGGGCCCAAGGTATTCTGCTAGTATATGACATCACAAATAAGTGGTCTTTTGATAGTATTGACAGGTGGCTTGAAGAAGTTGAAAAGCATGCACCCGGTGTCCCAAAAGTTCTGGTCGGGAATAGGTTACACCTCGCTTTCAAACGGCAAGTACAAGAACGGGATGCTGAAATGTATGCTGCAAAGAATCACATGGCTTTCTTCGAAGTGAGTCCGTTGTGCGACTTCAACATCAGGGAAAGTTTTTGTGAACTCTCACGGATGGCCCTTCACAGAAACGGCATGGAGAGACTCTGGAGGAGCAATAAAGTACTCAGTCTACAAGAGCTGTGCTGTCGTGCAATAGTAGCGCGGACATCCGTGTACGGTCTCGAAAGGCTTCCATTACCGAGCGCCCTGAAGTCCCATCTCAAGTCGTACGCGATATCGGCCGCTCCGAGCCGGCACCGAGCCCGCACGACCAAACACACGCATCATACAAGACTCAACTGCACCGGTCGTAACTCATGTACGATTGCCTGA
Protein
MTMFPASGSAAVPQQTINGQVVQNGSTTLPRSHHQRTGRPPAAPKSYDYLLKVLLVGDSDVGKQEILQDLEDGSADSPFCSGSGYKTTTILLDGKRVKLQLWDTSGQGRFCTIIRSYSRGAQGILLVYDITNKWSFDSIDRWLEEVEKHAPGVPKVLVGNRLHLAFKRQVQERDAEMYAAKNHMAFFEVSPLCDFNIRESFCELSRMALHRNGMERLWRSNKVLSLQELCCRAIVARTSVYGLERLPLPSALKSHLKSYAISAAPSRHRARTTKHTHHTRLNCTGRNSCTIA
Summary
Description
Interconversion of serine and glycine.
Catalytic Activity
(6R)-5,10-methylene-5,6,7,8-tetrahydrofolate + glycine + H2O = (6S)-5,6,7,8-tetrahydrofolate + L-serine
Cofactor
pyridoxal 5'-phosphate
Similarity
Belongs to the SHMT family.
Uniprot
A0A3S2LIX9
A0A194PHY6
A0A212FB49
E2GHU1
A0A194QYV9
H9JBX6
+ More
A0A212FDL8 A0A2J7REW5 A0A067RFT5 K7J5L0 A0A310S9F8 A0A2A3E109 A0A088AQW8 A0A0J7L2Q8 E1ZW03 A0A195DJQ1 A0A3L8DUB0 A0A195C9H2 F4WXC0 A0A158NU09 A0A154P0H1 A0A0C9PX33 A0A0M9AC19 A0A0A9YKY4 T1I6E5 A0A0K8TF99 A0A2P8YVP2 A0A0T6AWT5 A0A0L7R4W6 A0A151I4N5 A0A224XTD0 A0A023F5T0 A0A069DS17 E2BKI8 A0A1B6CV49 A0A1Y1M859 A0A195FJ48 N6TPL1 A0A2H1WNR7 D6WYZ4 A0A1W4X0U1 A0A1S3CZW3 A0A1B6L016 V5G8Z0 A0A0K8TNN2 D3TNL9 A0A1B0AMY0 A0A1B6M3C0 U5ETY4 A0A1A9WFW2 A0A1B6FCS8 A0A1B6IF75 A0A1I8NUS3 U4U098 A0A1B0CJM8 A0A0P4VHD7 A0A0V0GE20 Q171G7 B4N2F7 A0A2S2PX09 A0A182G7W9 A0A2H8TJR0 B4NTP8 B4IGF9 J9JV74 B4Q1M5 A0A0Q5TJA8 A0A1W4V2W8 A0A1Q3EYS6 B0XDP4 A0A0P8ZSG9 A0A182PWD1 A0A1L8DV32 A0A182K5D5 Q95RQ2 E0VST9 A0A182XXW4 Q9VYM8 A0A182N056 A0A182JLU6 A0A182QXV3 Q7Q4W5 A0A182HMN9 A0A182RMU7 A0A182SH89 A0A182LYF8 A0A182LDE9 A0A182W5E9 A0A2M3Z9N1 A0A2M4BX07 W5JID7 A0A1B0D222 A0A182FMN5 A0A2M4ATH9 A0A0A1XLC6 A0A0A1WR71 A0A0P4WIA7 A0A0A0PV43 A0A0K8W499
A0A212FDL8 A0A2J7REW5 A0A067RFT5 K7J5L0 A0A310S9F8 A0A2A3E109 A0A088AQW8 A0A0J7L2Q8 E1ZW03 A0A195DJQ1 A0A3L8DUB0 A0A195C9H2 F4WXC0 A0A158NU09 A0A154P0H1 A0A0C9PX33 A0A0M9AC19 A0A0A9YKY4 T1I6E5 A0A0K8TF99 A0A2P8YVP2 A0A0T6AWT5 A0A0L7R4W6 A0A151I4N5 A0A224XTD0 A0A023F5T0 A0A069DS17 E2BKI8 A0A1B6CV49 A0A1Y1M859 A0A195FJ48 N6TPL1 A0A2H1WNR7 D6WYZ4 A0A1W4X0U1 A0A1S3CZW3 A0A1B6L016 V5G8Z0 A0A0K8TNN2 D3TNL9 A0A1B0AMY0 A0A1B6M3C0 U5ETY4 A0A1A9WFW2 A0A1B6FCS8 A0A1B6IF75 A0A1I8NUS3 U4U098 A0A1B0CJM8 A0A0P4VHD7 A0A0V0GE20 Q171G7 B4N2F7 A0A2S2PX09 A0A182G7W9 A0A2H8TJR0 B4NTP8 B4IGF9 J9JV74 B4Q1M5 A0A0Q5TJA8 A0A1W4V2W8 A0A1Q3EYS6 B0XDP4 A0A0P8ZSG9 A0A182PWD1 A0A1L8DV32 A0A182K5D5 Q95RQ2 E0VST9 A0A182XXW4 Q9VYM8 A0A182N056 A0A182JLU6 A0A182QXV3 Q7Q4W5 A0A182HMN9 A0A182RMU7 A0A182SH89 A0A182LYF8 A0A182LDE9 A0A182W5E9 A0A2M3Z9N1 A0A2M4BX07 W5JID7 A0A1B0D222 A0A182FMN5 A0A2M4ATH9 A0A0A1XLC6 A0A0A1WR71 A0A0P4WIA7 A0A0A0PV43 A0A0K8W499
EC Number
2.1.2.1
Pubmed
26354079
22118469
19121390
24845553
20075255
20798317
+ More
30249741 21719571 21347285 25401762 26823975 29403074 25474469 26334808 28004739 23537049 18362917 19820115 26369729 20353571 27129103 17510324 17994087 26483478 17550304 20566863 25244985 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12364791 14747013 17210077 20966253 20920257 23761445 25830018
30249741 21719571 21347285 25401762 26823975 29403074 25474469 26334808 28004739 23537049 18362917 19820115 26369729 20353571 27129103 17510324 17994087 26483478 17550304 20566863 25244985 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12364791 14747013 17210077 20966253 20920257 23761445 25830018
EMBL
RSAL01000097
RVE47672.1
KQ459603
KPI93031.1
AGBW02009379
OWR50961.1
+ More
HM355929 ADN84947.1 KQ461137 KPJ08781.1 BABH01014822 AGBW02009020 OWR51846.1 NEVH01004429 PNF39378.1 KK852663 KDR19055.1 KQ762903 OAD55280.1 KZ288467 PBC25387.1 LBMM01001085 KMQ96953.1 GL434710 EFN74635.1 KQ980794 KYN13047.1 QOIP01000004 RLU24027.1 KQ978143 KYM96763.1 GL888423 EGI61169.1 ADTU01026236 KQ434782 KZC04640.1 GBYB01000996 GBYB01006063 JAG70763.1 JAG75830.1 KQ435698 KOX80756.1 GBHO01011841 GDHC01015806 GDHC01011772 JAG31763.1 JAQ02823.1 JAQ06857.1 ACPB03001813 GBRD01001549 JAG64272.1 PYGN01000334 PSN48322.1 LJIG01022721 KRT79063.1 KQ414657 KOC65859.1 KQ976445 KYM86066.1 GFTR01005063 JAW11363.1 GBBI01002177 JAC16535.1 GBGD01002154 JAC86735.1 GL448810 EFN83775.1 GEDC01019932 JAS17366.1 GEZM01038173 JAV81741.1 KQ981522 KYN40685.1 APGK01056846 KB741277 ENN71165.1 ODYU01009952 SOQ54713.1 KQ971363 EFA09306.2 GEBQ01022924 JAT17053.1 GALX01001861 JAB66605.1 GDAI01001872 JAI15731.1 EZ423021 ADD19297.1 JXJN01000640 GEBQ01009593 JAT30384.1 GANO01001711 JAB58160.1 GECZ01021757 JAS48012.1 GECU01022156 JAS85550.1 KB630186 KB632028 KB632155 ERL83460.1 ERL88124.1 ERL89222.1 AJWK01014755 GDKW01003559 JAI53036.1 GECL01000503 JAP05621.1 CH477456 EAT40624.1 CH963925 EDW78546.1 GGMS01000701 MBY69904.1 JXUM01046914 KQ561496 KXJ78387.1 GFXV01002474 MBW14279.1 CH983255 EDX16317.1 CH480836 EDW48909.1 ABLF02034859 CM000162 EDX01466.2 CH954180 KQS30443.1 GFDL01014657 JAV20388.1 DS232771 EDS45552.1 CH902630 KPU77439.1 GFDF01003798 JAV10286.1 AY061218 AAL28766.1 DS235756 EEB16445.1 AE014298 AAF48164.2 AHN59623.1 AXCN02001441 AAAB01008963 EAA12147.5 APCN01004851 AXCM01000689 GGFM01004482 MBW25233.1 GGFJ01008466 MBW57607.1 ADMH02001082 ETN64152.1 AJVK01002598 AJVK01002599 GGFK01010567 MBW43888.1 GBXI01002954 JAD11338.1 GBXI01013364 JAD00928.1 GDRN01035223 JAI67411.1 KF859983 AHJ90466.1 GDHF01006393 JAI45921.1
HM355929 ADN84947.1 KQ461137 KPJ08781.1 BABH01014822 AGBW02009020 OWR51846.1 NEVH01004429 PNF39378.1 KK852663 KDR19055.1 KQ762903 OAD55280.1 KZ288467 PBC25387.1 LBMM01001085 KMQ96953.1 GL434710 EFN74635.1 KQ980794 KYN13047.1 QOIP01000004 RLU24027.1 KQ978143 KYM96763.1 GL888423 EGI61169.1 ADTU01026236 KQ434782 KZC04640.1 GBYB01000996 GBYB01006063 JAG70763.1 JAG75830.1 KQ435698 KOX80756.1 GBHO01011841 GDHC01015806 GDHC01011772 JAG31763.1 JAQ02823.1 JAQ06857.1 ACPB03001813 GBRD01001549 JAG64272.1 PYGN01000334 PSN48322.1 LJIG01022721 KRT79063.1 KQ414657 KOC65859.1 KQ976445 KYM86066.1 GFTR01005063 JAW11363.1 GBBI01002177 JAC16535.1 GBGD01002154 JAC86735.1 GL448810 EFN83775.1 GEDC01019932 JAS17366.1 GEZM01038173 JAV81741.1 KQ981522 KYN40685.1 APGK01056846 KB741277 ENN71165.1 ODYU01009952 SOQ54713.1 KQ971363 EFA09306.2 GEBQ01022924 JAT17053.1 GALX01001861 JAB66605.1 GDAI01001872 JAI15731.1 EZ423021 ADD19297.1 JXJN01000640 GEBQ01009593 JAT30384.1 GANO01001711 JAB58160.1 GECZ01021757 JAS48012.1 GECU01022156 JAS85550.1 KB630186 KB632028 KB632155 ERL83460.1 ERL88124.1 ERL89222.1 AJWK01014755 GDKW01003559 JAI53036.1 GECL01000503 JAP05621.1 CH477456 EAT40624.1 CH963925 EDW78546.1 GGMS01000701 MBY69904.1 JXUM01046914 KQ561496 KXJ78387.1 GFXV01002474 MBW14279.1 CH983255 EDX16317.1 CH480836 EDW48909.1 ABLF02034859 CM000162 EDX01466.2 CH954180 KQS30443.1 GFDL01014657 JAV20388.1 DS232771 EDS45552.1 CH902630 KPU77439.1 GFDF01003798 JAV10286.1 AY061218 AAL28766.1 DS235756 EEB16445.1 AE014298 AAF48164.2 AHN59623.1 AXCN02001441 AAAB01008963 EAA12147.5 APCN01004851 AXCM01000689 GGFM01004482 MBW25233.1 GGFJ01008466 MBW57607.1 ADMH02001082 ETN64152.1 AJVK01002598 AJVK01002599 GGFK01010567 MBW43888.1 GBXI01002954 JAD11338.1 GBXI01013364 JAD00928.1 GDRN01035223 JAI67411.1 KF859983 AHJ90466.1 GDHF01006393 JAI45921.1
Proteomes
UP000283053
UP000053268
UP000007151
UP000053240
UP000005204
UP000235965
+ More
UP000027135 UP000002358 UP000242457 UP000005203 UP000036403 UP000000311 UP000078492 UP000279307 UP000078542 UP000007755 UP000005205 UP000076502 UP000053105 UP000015103 UP000245037 UP000053825 UP000078540 UP000008237 UP000078541 UP000019118 UP000007266 UP000192223 UP000079169 UP000092460 UP000091820 UP000095300 UP000030742 UP000092461 UP000008820 UP000007798 UP000069940 UP000249989 UP000000304 UP000001292 UP000007819 UP000002282 UP000008711 UP000192221 UP000002320 UP000007801 UP000075885 UP000075881 UP000009046 UP000076408 UP000000803 UP000075884 UP000075880 UP000075886 UP000007062 UP000075840 UP000075900 UP000075901 UP000075883 UP000075882 UP000075920 UP000000673 UP000092462 UP000069272
UP000027135 UP000002358 UP000242457 UP000005203 UP000036403 UP000000311 UP000078492 UP000279307 UP000078542 UP000007755 UP000005205 UP000076502 UP000053105 UP000015103 UP000245037 UP000053825 UP000078540 UP000008237 UP000078541 UP000019118 UP000007266 UP000192223 UP000079169 UP000092460 UP000091820 UP000095300 UP000030742 UP000092461 UP000008820 UP000007798 UP000069940 UP000249989 UP000000304 UP000001292 UP000007819 UP000002282 UP000008711 UP000192221 UP000002320 UP000007801 UP000075885 UP000075881 UP000009046 UP000076408 UP000000803 UP000075884 UP000075880 UP000075886 UP000007062 UP000075840 UP000075900 UP000075901 UP000075883 UP000075882 UP000075920 UP000000673 UP000092462 UP000069272
Interpro
IPR036036
SOCS_box-like_dom_sf
+ More
IPR001806 Small_GTPase
IPR027417 P-loop_NTPase
IPR001496 SOCS_box
IPR005225 Small_GTP-bd_dom
IPR015424 PyrdxlP-dep_Trfase
IPR015421 PyrdxlP-dep_Trfase_major
IPR015422 PyrdxlP-dep_Trfase_dom1
IPR039429 SHMT-like_dom
IPR019798 Ser_HO-MeTrfase_PLP_BS
IPR001085 Ser_HO-MeTrfase
IPR036322 WD40_repeat_dom_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR001806 Small_GTPase
IPR027417 P-loop_NTPase
IPR001496 SOCS_box
IPR005225 Small_GTP-bd_dom
IPR015424 PyrdxlP-dep_Trfase
IPR015421 PyrdxlP-dep_Trfase_major
IPR015422 PyrdxlP-dep_Trfase_dom1
IPR039429 SHMT-like_dom
IPR019798 Ser_HO-MeTrfase_PLP_BS
IPR001085 Ser_HO-MeTrfase
IPR036322 WD40_repeat_dom_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR015943 WD40/YVTN_repeat-like_dom_sf
Gene 3D
CDD
ProteinModelPortal
A0A3S2LIX9
A0A194PHY6
A0A212FB49
E2GHU1
A0A194QYV9
H9JBX6
+ More
A0A212FDL8 A0A2J7REW5 A0A067RFT5 K7J5L0 A0A310S9F8 A0A2A3E109 A0A088AQW8 A0A0J7L2Q8 E1ZW03 A0A195DJQ1 A0A3L8DUB0 A0A195C9H2 F4WXC0 A0A158NU09 A0A154P0H1 A0A0C9PX33 A0A0M9AC19 A0A0A9YKY4 T1I6E5 A0A0K8TF99 A0A2P8YVP2 A0A0T6AWT5 A0A0L7R4W6 A0A151I4N5 A0A224XTD0 A0A023F5T0 A0A069DS17 E2BKI8 A0A1B6CV49 A0A1Y1M859 A0A195FJ48 N6TPL1 A0A2H1WNR7 D6WYZ4 A0A1W4X0U1 A0A1S3CZW3 A0A1B6L016 V5G8Z0 A0A0K8TNN2 D3TNL9 A0A1B0AMY0 A0A1B6M3C0 U5ETY4 A0A1A9WFW2 A0A1B6FCS8 A0A1B6IF75 A0A1I8NUS3 U4U098 A0A1B0CJM8 A0A0P4VHD7 A0A0V0GE20 Q171G7 B4N2F7 A0A2S2PX09 A0A182G7W9 A0A2H8TJR0 B4NTP8 B4IGF9 J9JV74 B4Q1M5 A0A0Q5TJA8 A0A1W4V2W8 A0A1Q3EYS6 B0XDP4 A0A0P8ZSG9 A0A182PWD1 A0A1L8DV32 A0A182K5D5 Q95RQ2 E0VST9 A0A182XXW4 Q9VYM8 A0A182N056 A0A182JLU6 A0A182QXV3 Q7Q4W5 A0A182HMN9 A0A182RMU7 A0A182SH89 A0A182LYF8 A0A182LDE9 A0A182W5E9 A0A2M3Z9N1 A0A2M4BX07 W5JID7 A0A1B0D222 A0A182FMN5 A0A2M4ATH9 A0A0A1XLC6 A0A0A1WR71 A0A0P4WIA7 A0A0A0PV43 A0A0K8W499
A0A212FDL8 A0A2J7REW5 A0A067RFT5 K7J5L0 A0A310S9F8 A0A2A3E109 A0A088AQW8 A0A0J7L2Q8 E1ZW03 A0A195DJQ1 A0A3L8DUB0 A0A195C9H2 F4WXC0 A0A158NU09 A0A154P0H1 A0A0C9PX33 A0A0M9AC19 A0A0A9YKY4 T1I6E5 A0A0K8TF99 A0A2P8YVP2 A0A0T6AWT5 A0A0L7R4W6 A0A151I4N5 A0A224XTD0 A0A023F5T0 A0A069DS17 E2BKI8 A0A1B6CV49 A0A1Y1M859 A0A195FJ48 N6TPL1 A0A2H1WNR7 D6WYZ4 A0A1W4X0U1 A0A1S3CZW3 A0A1B6L016 V5G8Z0 A0A0K8TNN2 D3TNL9 A0A1B0AMY0 A0A1B6M3C0 U5ETY4 A0A1A9WFW2 A0A1B6FCS8 A0A1B6IF75 A0A1I8NUS3 U4U098 A0A1B0CJM8 A0A0P4VHD7 A0A0V0GE20 Q171G7 B4N2F7 A0A2S2PX09 A0A182G7W9 A0A2H8TJR0 B4NTP8 B4IGF9 J9JV74 B4Q1M5 A0A0Q5TJA8 A0A1W4V2W8 A0A1Q3EYS6 B0XDP4 A0A0P8ZSG9 A0A182PWD1 A0A1L8DV32 A0A182K5D5 Q95RQ2 E0VST9 A0A182XXW4 Q9VYM8 A0A182N056 A0A182JLU6 A0A182QXV3 Q7Q4W5 A0A182HMN9 A0A182RMU7 A0A182SH89 A0A182LYF8 A0A182LDE9 A0A182W5E9 A0A2M3Z9N1 A0A2M4BX07 W5JID7 A0A1B0D222 A0A182FMN5 A0A2M4ATH9 A0A0A1XLC6 A0A0A1WR71 A0A0P4WIA7 A0A0A0PV43 A0A0K8W499
PDB
2FU5
E-value=4.74791e-36,
Score=377
Ontologies
GO
PANTHER
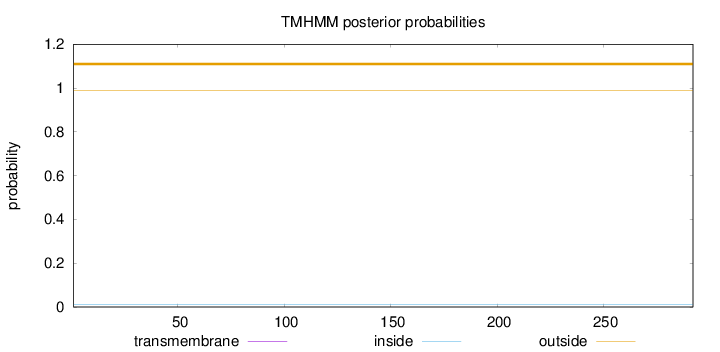
Topology
Length:
292
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00013
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01059
outside
1 - 292
Population Genetic Test Statistics
Pi
150.573365
Theta
205.997734
Tajima's D
-1.29499
CLR
0.542773
CSRT
0.0905954702264887
Interpretation
Possibly Positive selection
