Gene
KWMTBOMO10418
Pre Gene Modal
BGIBMGA007080
Annotation
PREDICTED:_rRNA_methyltransferase_2?_mitochondrial_[Papilio_polytes]
Full name
rRNA methyltransferase 2, mitochondrial
Alternative Name
rRNA (uridine-2'-O)-methyltransferase
Location in the cell
Mitochondrial Reliability : 2.398
Sequence
CDS
ATGCTTTGTTCGCATCGAGAAGTGACGCATATCAAATTGAATTACACGAGAATGTGCTTCTTAGTGAATTGTCTAAAAAGATTTAAAAGTTCACAAGAATGGCTGAGTCGTCAAAAGGCTGATCCTTATGTAGAGAAAGCTAAGATATACAACTATAGATGTCGCAGTGCCTTCAAATTATTGGAGATGAATGAAAGAACCAATATATTGAGACCTGGCCTAGCTGTTATTGATCTTGGAGCATGTCCCGGTTCTTGGACGCAAGTGGCGGTTCAGAAAACAAATGCGGATGGTGCAGATCCTAGTAAGCCCAAGGGAACAGTCTTGTCAATTGATAAACTTCAGATTTTTCCTATTCAGGGAGCAACAATACTAAATAACATGGATTTCTCTACAATTGAAGCACATGACAAAGTTGTAGAAGTTTTAAATGGCCAACAAGTTGATCTAGTTCTCTCTGACATGGCTCCCAGTGCAACAGGTGTCCGGGAATTGGATAAAGATAGAATAATTGGTCTCTGTTACATGGCTATCAGATTTGCAGCACTTGTGACAAAAAGAGAAGGAAATTTACTATTTAAAGTATGGGATGGAAAAGAAGTTCCTATACTAGAAATGGATTTAGCAAGGTTTTATAAGAATATAAAGATATTAAAGCCGATGGCAAGCAGGACTGAATCTTCAGAAAAGTTTATTTTAGCAAAGGGATTCAAGGGGATACAGAGACCTTTACAAAATGGCAGATGGGGTTGA
Protein
MLCSHREVTHIKLNYTRMCFLVNCLKRFKSSQEWLSRQKADPYVEKAKIYNYRCRSAFKLLEMNERTNILRPGLAVIDLGACPGSWTQVAVQKTNADGADPSKPKGTVLSIDKLQIFPIQGATILNNMDFSTIEAHDKVVEVLNGQQVDLVLSDMAPSATGVRELDKDRIIGLCYMAIRFAALVTKREGNLLFKVWDGKEVPILEMDLARFYKNIKILKPMASRTESSEKFILAKGFKGIQRPLQNGRWG
Summary
Description
S-adenosyl-L-methionine-dependent 2'-O-ribose methyltransferase that catalyzes the formation of 2'-O-methyluridine at position 1579 (Um1579) in the mitochondrial large subunit ribosomal RNA (mtLSU rRNA), a universally conserved modification in the peptidyl transferase domain of the mtLSU rRNA.
Catalytic Activity
a uridine in rRNA + S-adenosyl-L-methionine = a 2'-O-methyluridine in rRNA + H(+) + S-adenosyl-L-homocysteine
Similarity
Belongs to the class I-like SAM-binding methyltransferase superfamily. RNA methyltransferase RlmE family.
Keywords
Complete proteome
Methyltransferase
Mitochondrion
Reference proteome
rRNA processing
S-adenosyl-L-methionine
Transferase
Transit peptide
Feature
chain rRNA methyltransferase 2, mitochondrial
Uniprot
H9JC35
A0A0L7KUK3
A0A194PJ07
A0A194QTD1
A0A212F0S3
S4P8C4
+ More
A0A1Q3FGX1 B0W330 Q16QF4 A0A2A4J0J3 A0A1I8NE31 A0A232FKL9 A0A084VBR9 K7IZI4 A0A2P8YY69 A0A182YSD8 B3LXS3 A0A2J7QN05 A0A2R7VZL4 W5JUN1 A0A1B0BLV9 A0A1A9Y8J1 E9IQA8 A0A026WVI2 A0A182XJ46 A0A1A9VAE4 A0A154PRT1 A0A0L0CHB3 A0A182L7B0 D6WYW4 T1DU58 Q7QB53 A0A182QUS8 A0A182I7R5 A0A182P6R9 A0A1B0FQT1 A0A0C9R8E9 A0A182VJ31 A0A182W1G8 A0A182JTD0 A0A1Y1LBB9 A0A182NLC9 A0A0M4ENT7 A0A1I8PYJ7 F5HL46 A0A182LS23 A0A0J7KZ80 A0A182U4D2 E2ANP9 A0A3B0JNI6 A0A182RYP5 B4IV03 B4GMN5 B3P080 B4PNK4 Q9VDT6 A0A1W4VXH1 B4QST8 A0A0N0BCN8 A0A195CYH7 B4JT12 Q294S6 W8C1M3 A0A0L7QV70 B4IHR8 A0A0K8SBI6 B4K4S3 A0A2A3E7L8 B4NEY9 A0A146M257 X1WLQ5 A0A1S3D6B1 A0A1S4EFC6 A0A1A9WJ71 A0A2H8TU79 A0A0P4VHN9 R4FJT4 A0A0A9YYJ6 B4M4R8 A0A2S2P7K5 A0A195BU54 A0A158NKP8 A0A195EJ81 A0A034VKW6 A0A195F4R1 F4WD06 A0A0K8VUB2 A0A023F7T0 A0A310SGG9 A0A088AEE5 A0A151X8F0 N6UGP9 A0A0V0GB01 A0A1B6CF18 A0A182JGN7 A0A1W4XS80 E2BQR2 A0A2S2R587
A0A1Q3FGX1 B0W330 Q16QF4 A0A2A4J0J3 A0A1I8NE31 A0A232FKL9 A0A084VBR9 K7IZI4 A0A2P8YY69 A0A182YSD8 B3LXS3 A0A2J7QN05 A0A2R7VZL4 W5JUN1 A0A1B0BLV9 A0A1A9Y8J1 E9IQA8 A0A026WVI2 A0A182XJ46 A0A1A9VAE4 A0A154PRT1 A0A0L0CHB3 A0A182L7B0 D6WYW4 T1DU58 Q7QB53 A0A182QUS8 A0A182I7R5 A0A182P6R9 A0A1B0FQT1 A0A0C9R8E9 A0A182VJ31 A0A182W1G8 A0A182JTD0 A0A1Y1LBB9 A0A182NLC9 A0A0M4ENT7 A0A1I8PYJ7 F5HL46 A0A182LS23 A0A0J7KZ80 A0A182U4D2 E2ANP9 A0A3B0JNI6 A0A182RYP5 B4IV03 B4GMN5 B3P080 B4PNK4 Q9VDT6 A0A1W4VXH1 B4QST8 A0A0N0BCN8 A0A195CYH7 B4JT12 Q294S6 W8C1M3 A0A0L7QV70 B4IHR8 A0A0K8SBI6 B4K4S3 A0A2A3E7L8 B4NEY9 A0A146M257 X1WLQ5 A0A1S3D6B1 A0A1S4EFC6 A0A1A9WJ71 A0A2H8TU79 A0A0P4VHN9 R4FJT4 A0A0A9YYJ6 B4M4R8 A0A2S2P7K5 A0A195BU54 A0A158NKP8 A0A195EJ81 A0A034VKW6 A0A195F4R1 F4WD06 A0A0K8VUB2 A0A023F7T0 A0A310SGG9 A0A088AEE5 A0A151X8F0 N6UGP9 A0A0V0GB01 A0A1B6CF18 A0A182JGN7 A0A1W4XS80 E2BQR2 A0A2S2R587
EC Number
2.1.1.-
Pubmed
19121390
26227816
26354079
22118469
23622113
17510324
+ More
25315136 28648823 24438588 20075255 29403074 25244985 17994087 20920257 23761445 21282665 24508170 30249741 26108605 20966253 18362917 19820115 12364791 14747013 17210077 28004739 20798317 17550304 10731132 12537572 12537569 25009282 15632085 24495485 26823975 27129103 25401762 21347285 25348373 21719571 25474469 23537049
25315136 28648823 24438588 20075255 29403074 25244985 17994087 20920257 23761445 21282665 24508170 30249741 26108605 20966253 18362917 19820115 12364791 14747013 17210077 28004739 20798317 17550304 10731132 12537572 12537569 25009282 15632085 24495485 26823975 27129103 25401762 21347285 25348373 21719571 25474469 23537049
EMBL
BABH01014821
JTDY01005699
KOB66731.1
KQ459603
KPI93033.1
KQ461137
+ More
KPJ08783.1 AGBW02011057 OWR47301.1 GAIX01009565 JAA82995.1 GFDL01008239 JAV26806.1 DS231830 EDS30738.1 CH477750 EAT36622.1 NWSH01004560 PCG64932.1 NNAY01000090 OXU31080.1 ATLV01009141 KE524507 KFB35413.1 PYGN01000290 PSN49184.1 CH902617 EDV41730.1 NEVH01013201 PNF29971.1 KK854104 PTY11355.1 ADMH02000319 ETN66983.1 JXJN01016583 JXJN01016584 JXJN01016585 JXJN01016586 GL764688 EFZ17244.1 KK107087 QOIP01000001 EZA59998.1 RLU27177.1 KQ435012 KZC13820.1 JRES01000405 KNC31640.1 KQ971363 EFA09016.2 GAMD01000444 JAB01147.1 AAAB01008880 EAA08541.4 AXCN02000121 APCN01002549 CCAG010014772 GBYB01004320 JAG74087.1 GEZM01062524 GEZM01062522 GEZM01062520 GEZM01062519 GEZM01062517 GEZM01062516 JAV69640.1 CP012526 ALC47340.1 EGK97007.1 AXCM01003760 LBMM01001891 KMQ95598.1 GL441295 EFN64935.1 OUUW01000008 SPP83705.1 CH892421 EDX00217.1 CH479185 EDW38109.1 CH954181 EDV48454.1 CM000160 EDW96067.1 AE014297 AY071418 CM000364 EDX12298.1 KQ435908 KOX68972.1 KQ977110 KYN05725.1 CH916373 EDV94902.1 CM000070 EAL28888.3 GAMC01010991 JAB95564.1 KQ414734 KOC62341.1 CH480840 EDW49435.1 GBRD01015164 GDHC01006439 JAG50662.1 JAQ12190.1 CH933806 EDW16076.2 KZ288355 PBC27216.1 CH964251 EDW83364.1 GDHC01004758 JAQ13871.1 ABLF02003617 GFXV01005991 MBW17796.1 GDKW01002504 JAI54091.1 ACPB03000203 GAHY01002148 JAA75362.1 GBHO01008994 JAG34610.1 CH940652 EDW59629.1 GGMR01012766 MBY25385.1 KQ976403 KYM92112.1 ADTU01002112 KQ978801 KYN28320.1 GAKP01016779 JAC42173.1 KQ981805 KYN35570.1 GL888077 EGI67913.1 GDHF01009866 JAI42448.1 GBBI01001415 JAC17297.1 KQ770367 OAD52660.1 KQ982431 KYQ56558.1 APGK01036043 KB740932 KB632408 ENN77827.1 ERL95122.1 GECL01001104 JAP05020.1 GEDC01025254 JAS12044.1 GL449798 EFN81981.1 GGMS01016004 MBY85207.1
KPJ08783.1 AGBW02011057 OWR47301.1 GAIX01009565 JAA82995.1 GFDL01008239 JAV26806.1 DS231830 EDS30738.1 CH477750 EAT36622.1 NWSH01004560 PCG64932.1 NNAY01000090 OXU31080.1 ATLV01009141 KE524507 KFB35413.1 PYGN01000290 PSN49184.1 CH902617 EDV41730.1 NEVH01013201 PNF29971.1 KK854104 PTY11355.1 ADMH02000319 ETN66983.1 JXJN01016583 JXJN01016584 JXJN01016585 JXJN01016586 GL764688 EFZ17244.1 KK107087 QOIP01000001 EZA59998.1 RLU27177.1 KQ435012 KZC13820.1 JRES01000405 KNC31640.1 KQ971363 EFA09016.2 GAMD01000444 JAB01147.1 AAAB01008880 EAA08541.4 AXCN02000121 APCN01002549 CCAG010014772 GBYB01004320 JAG74087.1 GEZM01062524 GEZM01062522 GEZM01062520 GEZM01062519 GEZM01062517 GEZM01062516 JAV69640.1 CP012526 ALC47340.1 EGK97007.1 AXCM01003760 LBMM01001891 KMQ95598.1 GL441295 EFN64935.1 OUUW01000008 SPP83705.1 CH892421 EDX00217.1 CH479185 EDW38109.1 CH954181 EDV48454.1 CM000160 EDW96067.1 AE014297 AY071418 CM000364 EDX12298.1 KQ435908 KOX68972.1 KQ977110 KYN05725.1 CH916373 EDV94902.1 CM000070 EAL28888.3 GAMC01010991 JAB95564.1 KQ414734 KOC62341.1 CH480840 EDW49435.1 GBRD01015164 GDHC01006439 JAG50662.1 JAQ12190.1 CH933806 EDW16076.2 KZ288355 PBC27216.1 CH964251 EDW83364.1 GDHC01004758 JAQ13871.1 ABLF02003617 GFXV01005991 MBW17796.1 GDKW01002504 JAI54091.1 ACPB03000203 GAHY01002148 JAA75362.1 GBHO01008994 JAG34610.1 CH940652 EDW59629.1 GGMR01012766 MBY25385.1 KQ976403 KYM92112.1 ADTU01002112 KQ978801 KYN28320.1 GAKP01016779 JAC42173.1 KQ981805 KYN35570.1 GL888077 EGI67913.1 GDHF01009866 JAI42448.1 GBBI01001415 JAC17297.1 KQ770367 OAD52660.1 KQ982431 KYQ56558.1 APGK01036043 KB740932 KB632408 ENN77827.1 ERL95122.1 GECL01001104 JAP05020.1 GEDC01025254 JAS12044.1 GL449798 EFN81981.1 GGMS01016004 MBY85207.1
Proteomes
UP000005204
UP000037510
UP000053268
UP000053240
UP000007151
UP000002320
+ More
UP000008820 UP000218220 UP000095301 UP000215335 UP000030765 UP000002358 UP000245037 UP000076408 UP000007801 UP000235965 UP000000673 UP000092460 UP000092443 UP000053097 UP000279307 UP000076407 UP000078200 UP000076502 UP000037069 UP000075882 UP000007266 UP000007062 UP000075886 UP000075840 UP000075885 UP000092444 UP000075903 UP000075920 UP000075881 UP000075884 UP000092553 UP000095300 UP000075883 UP000036403 UP000075902 UP000000311 UP000268350 UP000075900 UP000002282 UP000008744 UP000008711 UP000000803 UP000192221 UP000000304 UP000053105 UP000078542 UP000001070 UP000001819 UP000053825 UP000001292 UP000009192 UP000242457 UP000007798 UP000007819 UP000079169 UP000091820 UP000015103 UP000008792 UP000078540 UP000005205 UP000078492 UP000078541 UP000007755 UP000005203 UP000075809 UP000019118 UP000030742 UP000075880 UP000192223 UP000008237
UP000008820 UP000218220 UP000095301 UP000215335 UP000030765 UP000002358 UP000245037 UP000076408 UP000007801 UP000235965 UP000000673 UP000092460 UP000092443 UP000053097 UP000279307 UP000076407 UP000078200 UP000076502 UP000037069 UP000075882 UP000007266 UP000007062 UP000075886 UP000075840 UP000075885 UP000092444 UP000075903 UP000075920 UP000075881 UP000075884 UP000092553 UP000095300 UP000075883 UP000036403 UP000075902 UP000000311 UP000268350 UP000075900 UP000002282 UP000008744 UP000008711 UP000000803 UP000192221 UP000000304 UP000053105 UP000078542 UP000001070 UP000001819 UP000053825 UP000001292 UP000009192 UP000242457 UP000007798 UP000007819 UP000079169 UP000091820 UP000015103 UP000008792 UP000078540 UP000005205 UP000078492 UP000078541 UP000007755 UP000005203 UP000075809 UP000019118 UP000030742 UP000075880 UP000192223 UP000008237
Pfam
PF01728 FtsJ
Interpro
SUPFAM
SSF53335
SSF53335
ProteinModelPortal
H9JC35
A0A0L7KUK3
A0A194PJ07
A0A194QTD1
A0A212F0S3
S4P8C4
+ More
A0A1Q3FGX1 B0W330 Q16QF4 A0A2A4J0J3 A0A1I8NE31 A0A232FKL9 A0A084VBR9 K7IZI4 A0A2P8YY69 A0A182YSD8 B3LXS3 A0A2J7QN05 A0A2R7VZL4 W5JUN1 A0A1B0BLV9 A0A1A9Y8J1 E9IQA8 A0A026WVI2 A0A182XJ46 A0A1A9VAE4 A0A154PRT1 A0A0L0CHB3 A0A182L7B0 D6WYW4 T1DU58 Q7QB53 A0A182QUS8 A0A182I7R5 A0A182P6R9 A0A1B0FQT1 A0A0C9R8E9 A0A182VJ31 A0A182W1G8 A0A182JTD0 A0A1Y1LBB9 A0A182NLC9 A0A0M4ENT7 A0A1I8PYJ7 F5HL46 A0A182LS23 A0A0J7KZ80 A0A182U4D2 E2ANP9 A0A3B0JNI6 A0A182RYP5 B4IV03 B4GMN5 B3P080 B4PNK4 Q9VDT6 A0A1W4VXH1 B4QST8 A0A0N0BCN8 A0A195CYH7 B4JT12 Q294S6 W8C1M3 A0A0L7QV70 B4IHR8 A0A0K8SBI6 B4K4S3 A0A2A3E7L8 B4NEY9 A0A146M257 X1WLQ5 A0A1S3D6B1 A0A1S4EFC6 A0A1A9WJ71 A0A2H8TU79 A0A0P4VHN9 R4FJT4 A0A0A9YYJ6 B4M4R8 A0A2S2P7K5 A0A195BU54 A0A158NKP8 A0A195EJ81 A0A034VKW6 A0A195F4R1 F4WD06 A0A0K8VUB2 A0A023F7T0 A0A310SGG9 A0A088AEE5 A0A151X8F0 N6UGP9 A0A0V0GB01 A0A1B6CF18 A0A182JGN7 A0A1W4XS80 E2BQR2 A0A2S2R587
A0A1Q3FGX1 B0W330 Q16QF4 A0A2A4J0J3 A0A1I8NE31 A0A232FKL9 A0A084VBR9 K7IZI4 A0A2P8YY69 A0A182YSD8 B3LXS3 A0A2J7QN05 A0A2R7VZL4 W5JUN1 A0A1B0BLV9 A0A1A9Y8J1 E9IQA8 A0A026WVI2 A0A182XJ46 A0A1A9VAE4 A0A154PRT1 A0A0L0CHB3 A0A182L7B0 D6WYW4 T1DU58 Q7QB53 A0A182QUS8 A0A182I7R5 A0A182P6R9 A0A1B0FQT1 A0A0C9R8E9 A0A182VJ31 A0A182W1G8 A0A182JTD0 A0A1Y1LBB9 A0A182NLC9 A0A0M4ENT7 A0A1I8PYJ7 F5HL46 A0A182LS23 A0A0J7KZ80 A0A182U4D2 E2ANP9 A0A3B0JNI6 A0A182RYP5 B4IV03 B4GMN5 B3P080 B4PNK4 Q9VDT6 A0A1W4VXH1 B4QST8 A0A0N0BCN8 A0A195CYH7 B4JT12 Q294S6 W8C1M3 A0A0L7QV70 B4IHR8 A0A0K8SBI6 B4K4S3 A0A2A3E7L8 B4NEY9 A0A146M257 X1WLQ5 A0A1S3D6B1 A0A1S4EFC6 A0A1A9WJ71 A0A2H8TU79 A0A0P4VHN9 R4FJT4 A0A0A9YYJ6 B4M4R8 A0A2S2P7K5 A0A195BU54 A0A158NKP8 A0A195EJ81 A0A034VKW6 A0A195F4R1 F4WD06 A0A0K8VUB2 A0A023F7T0 A0A310SGG9 A0A088AEE5 A0A151X8F0 N6UGP9 A0A0V0GB01 A0A1B6CF18 A0A182JGN7 A0A1W4XS80 E2BQR2 A0A2S2R587
PDB
2NYU
E-value=2.20936e-50,
Score=500
Ontologies
GO
Topology
Subcellular location
Mitochondrion
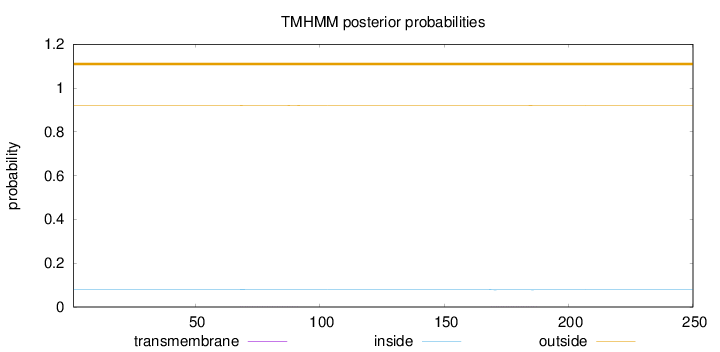
Length:
250
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03791
Exp number, first 60 AAs:
0.0005
Total prob of N-in:
0.08005
outside
1 - 250
Population Genetic Test Statistics
Pi
346.640649
Theta
235.958352
Tajima's D
1.46771
CLR
0.022907
CSRT
0.779861006949653
Interpretation
Possibly Positive selection
