Pre Gene Modal
BGIBMGA007030
Annotation
PREDICTED:_spermatogenesis-associated_protein_20-like_isoform_X2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.886
Sequence
CDS
ATGCTATCCTCAAGTTCAAAAACTCAATCAGTTGCAGATAAACCTAAATATACAAATGGTTTAATCAATGAAAAATCGCCCTACCTACTTCAACATGCTCACAATCCTGTGCAATGGTATCCTTGGGGTCCAGAAGCTATAGAAAAGGCAAAGGTGCAAAAGAAATTGATATTCTTATCAGTAGGCTATTCTACATGTCACTGGTGTCATGTTATGGAAAAAGAGTCCTTTGAGAATGAAAATGTTGCAAAAATTATGAATGATCATTATATTAACATCAAAGTTGACAGAGAAGAGAGACCTGATATTGATCTTGTTTATTTGTCTTTTGTCATGGCATCCACAGGAAATGAAAGCTGGCCAATGTCTGTCTTTTTAACTCCAGATCTTCAACCGGTCTCTGGAGGTACCTATTTCCCTCCAGAAGATCGCTGGGGGCATCTCGGTTTCAAAACAGCCCTTCTTACTTTAGCTAAGAAATGGAAAGAAAATCCACAACAGTGTCTTGAGGTCGGTCAACATGTAATTATCGCATTACAACAATTATCTGTTGATGACAATCTATCTACGGTAGTCTTAGGAGAGGGTATTTGGGACAAATGTGTTCAAAAATATATGTTGTTATATGAAACAGAGTTTGGTGGATTTGGAATGGCTCCTAAATTTCCACACGCATCAATTTTAAATTTCCATTTCCATTACTATGCTAGAGATAAATCAAACCCTGATGGAAAAAAATGTCTAGAAATGTGTTTGTACACTTTGACTAAGATCGCTAAAGGAGCTGTTCATGACCATGTATGCAGTGGATTTGCACGTTATTCAGATGGCAATGATTGGCATGTTCATCACTTTGAAAAGATGTTGTATGATCAAGCTCAGTTGGTAGTTGCCTACACTGATGCCTTCCTGGCAACTAAGGATGAATTTTACGCTGATATAGTTTATGACATAATCAAATATGTCAACAGAGATCTGAGACATGAATCTGGTGGATATTTCAGTGCTGAAGATTCAGACTCGTTTCCCACTTTTGAAACTACTTATAAAAAAGAAGGTGCCTTTTGTCTATGGGAATATAATGAACTAAAAGAACTACTAAATGATAAAAAGATAAATGATATATCTTACTTGGAAATTTTTTGTGAATATTTTTCTATTAATGAAGAAGGAAATATATCTCCGATTACTGACCTGCGCGGAGAACTAGTACACAAAAATGCCTTAATTATTTATGACAGTAAAGAGGAAATTATCCAAAAATTTAATTTAAGCACTGAAGAATTTAACCAAGTCATCAAAGACTGTATTAAAATTGTATATGATGCACGAAAAAATCGTCCCCGGCCAGACTTAGACACAAAAATTTTGTGTTCTTGGAATGGGCTAATGCTATCTGCATTGGCACAAGCTGGACAAGGTTTGGGAGAGAATAGTTTTGTTGATGATGCCATTAAGACTGCCTATTTTATAAAAAAATACCTATACGATGTAACCACGACCACTTTACTGCACTCCTGTTACAGAGATGACAATGGGGACATCTTAAAAATGAAAAATCCAATAAAAGCTTTTTTGGATGATTACGCTTTTCTTATCAAAGGACTTCTCGATTTGTACGAAGCATCGTTTGATCGGCGTTGGTTGAATTGGGCCCGTGAATTACAATTGAAGCAAAATGACATGTTCTGGGATTCAAAAGATGGTGGTTACTTTACGTGTTCAGCCGAAGATAAAACTATAGTGTTGAGACTTAAAGAAGATCACGATGGTGCCGAACCGTCGGGTAATAGTGTTTCGTGCCACAACCTGCTTCGGCTCGCGGCCCACGCCGACAAGAGTGATGAAGAAAATGGCAGTGAGAAGAAAAGAGATATGGCTAAAAAGATACTTGTGGCTTTTGCAAAACGTCTCAACGACTCGCCCACTGCCCTACCCGAAATGATGTCCGCATTGATGTTTTACAATGATTCTCCCACAAAGGTGCTGATCTCGGGGGGCCGCTCGGACCCGCGCACGCTGGAGATGGTGCGCGCGGTGCGGACGCGGCTGCTGCCGGGCCGCGTGCTGTGCGTGGCCGACCCCGCCGACACGCCCGCCGTGTTGCACCATGTGAAGCCTGCGAATGTAGGGCCGCGCGCGCACGTGTGCCGCCGCTACGCGTGTTCGCTTCCCCTCACAGACGTCAAACAACTCGAGAACTTGCTAGACGAAACACCCGCTGCTGTCGCCAAGAAATAG
Protein
MLSSSSKTQSVADKPKYTNGLINEKSPYLLQHAHNPVQWYPWGPEAIEKAKVQKKLIFLSVGYSTCHWCHVMEKESFENENVAKIMNDHYINIKVDREERPDIDLVYLSFVMASTGNESWPMSVFLTPDLQPVSGGTYFPPEDRWGHLGFKTALLTLAKKWKENPQQCLEVGQHVIIALQQLSVDDNLSTVVLGEGIWDKCVQKYMLLYETEFGGFGMAPKFPHASILNFHFHYYARDKSNPDGKKCLEMCLYTLTKIAKGAVHDHVCSGFARYSDGNDWHVHHFEKMLYDQAQLVVAYTDAFLATKDEFYADIVYDIIKYVNRDLRHESGGYFSAEDSDSFPTFETTYKKEGAFCLWEYNELKELLNDKKINDISYLEIFCEYFSINEEGNISPITDLRGELVHKNALIIYDSKEEIIQKFNLSTEEFNQVIKDCIKIVYDARKNRPRPDLDTKILCSWNGLMLSALAQAGQGLGENSFVDDAIKTAYFIKKYLYDVTTTTLLHSCYRDDNGDILKMKNPIKAFLDDYAFLIKGLLDLYEASFDRRWLNWARELQLKQNDMFWDSKDGGYFTCSAEDKTIVLRLKEDHDGAEPSGNSVSCHNLLRLAAHADKSDEENGSEKKRDMAKKILVAFAKRLNDSPTALPEMMSALMFYNDSPTKVLISGGRSDPRTLEMVRAVRTRLLPGRVLCVADPADTPAVLHHVKPANVGPRAHVCRRYACSLPLTDVKQLENLLDETPAAVAKK
Summary
Uniprot
H9JBY5
H9JBY2
A0A194PI78
A0A212FG87
A0A194R4Q1
A0A0L7KN86
+ More
A0A0L7LF60 D6WUV1 A0A1Y1KJS0 A0A2J7RRZ6 A0A067QYI6 A0A0T6B6Q9 A0A1B6EDT0 A0A1B6DA39 E2B1K9 K7IWR6 A0A2H8TMN0 C3Z6V7 J9JUE2 A0A2C9K6P6 A0A154P5R6 E2B587 A0A1W4WG18 E0VAZ1 A0A088ATM7 A0A026VZ87 A0A3L8DSQ5 A0A146KV46 A0A0P6E3M5 A0A0A9VSA9 U3JF44 A0A0P6JJ08 A0A0P6DTV0 A0A0P6D9B1 A0A0P5SL14 A0A0P5L037 A0A0P5NQP0 A0A0P5FPK1 A0A0P6CR19 A0A0P5IHA0 A0A0P5HHQ1 A0A0N8B7F1 A0A0P5S7K9 A0A0P5J486 A0A0P5G7Y3 A0A2A3E2K1 A0A1A8FLL3 A0A147A8I6 A0A3Q2Y1H6 A0A3Q2C6R7 A0A1A8CNB0 A0A1A8HUK2 A0A3P8QME6 A0A1A8NWB8 A0A3P8QKZ9 A0A1S3K5P5 A0A091I1Q1 A0A3Q2V915 A0A3Q2ZHB3 A0A0P6DIU8 A0A1A8A7P1 A0A0P5JKA5 A0A0P5KWY8 A0A091GZY1 A0A091NUU6 A0A1V4KW33 A0A0Q3SYV2 A0A091EEA5 A0A1A8PQY4 A0A1A7Y227 A0A3B4U3R2 H3CFR8 A0A093GA60 A0A3B3Y5S5 A0A096MGL3 A0A3B3W0G2 A0A3B4YEX7 A0A091VY85 A0A3P8WW56 M7B7C5 F1NIQ1 H2V487 A0A3Q2TSE6 A0A087R2X4 A0A3B4FBR3 A0A151N2U7 A0A151N2Y0 A0A3Q0SWV1 E9G951 A0A3B5AKG1 A0A2I4DBF1 K7FMX6 A0A3Q1IMD4 A0A1W5AW95 E7F7E2 A0A3P9MUN7 A0A091J299 A0A2A3E1C8 A0A1U7R9M8 A0A023F3M5
A0A0L7LF60 D6WUV1 A0A1Y1KJS0 A0A2J7RRZ6 A0A067QYI6 A0A0T6B6Q9 A0A1B6EDT0 A0A1B6DA39 E2B1K9 K7IWR6 A0A2H8TMN0 C3Z6V7 J9JUE2 A0A2C9K6P6 A0A154P5R6 E2B587 A0A1W4WG18 E0VAZ1 A0A088ATM7 A0A026VZ87 A0A3L8DSQ5 A0A146KV46 A0A0P6E3M5 A0A0A9VSA9 U3JF44 A0A0P6JJ08 A0A0P6DTV0 A0A0P6D9B1 A0A0P5SL14 A0A0P5L037 A0A0P5NQP0 A0A0P5FPK1 A0A0P6CR19 A0A0P5IHA0 A0A0P5HHQ1 A0A0N8B7F1 A0A0P5S7K9 A0A0P5J486 A0A0P5G7Y3 A0A2A3E2K1 A0A1A8FLL3 A0A147A8I6 A0A3Q2Y1H6 A0A3Q2C6R7 A0A1A8CNB0 A0A1A8HUK2 A0A3P8QME6 A0A1A8NWB8 A0A3P8QKZ9 A0A1S3K5P5 A0A091I1Q1 A0A3Q2V915 A0A3Q2ZHB3 A0A0P6DIU8 A0A1A8A7P1 A0A0P5JKA5 A0A0P5KWY8 A0A091GZY1 A0A091NUU6 A0A1V4KW33 A0A0Q3SYV2 A0A091EEA5 A0A1A8PQY4 A0A1A7Y227 A0A3B4U3R2 H3CFR8 A0A093GA60 A0A3B3Y5S5 A0A096MGL3 A0A3B3W0G2 A0A3B4YEX7 A0A091VY85 A0A3P8WW56 M7B7C5 F1NIQ1 H2V487 A0A3Q2TSE6 A0A087R2X4 A0A3B4FBR3 A0A151N2U7 A0A151N2Y0 A0A3Q0SWV1 E9G951 A0A3B5AKG1 A0A2I4DBF1 K7FMX6 A0A3Q1IMD4 A0A1W5AW95 E7F7E2 A0A3P9MUN7 A0A091J299 A0A2A3E1C8 A0A1U7R9M8 A0A023F3M5
Pubmed
EMBL
BABH01014791
BABH01014793
KQ459603
KPI93017.1
AGBW02008703
OWR52746.1
+ More
KQ460685 KPJ12793.1 JTDY01008121 KOB64727.1 JTDY01001371 KOB74045.1 KQ971363 EFA07789.1 GEZM01085937 JAV59875.1 NEVH01000597 PNF43606.1 KK852872 KDR14558.1 LJIG01009532 KRT82891.1 GEDC01001248 JAS36050.1 GEDC01023860 GEDC01021755 GEDC01018561 GEDC01014843 JAS13438.1 JAS15543.1 JAS18737.1 JAS22455.1 GL444953 EFN60365.1 AAZX01000604 GFXV01003484 MBW15289.1 GG666590 EEN51553.1 ABLF02026651 KQ434823 KZC07269.1 GL445739 EFN89140.1 DS235019 EEB10547.1 KK107570 EZA48786.1 QOIP01000005 RLU22909.1 GDHC01018461 GDHC01018295 GDHC01008407 GDHC01004818 JAQ00168.1 JAQ00334.1 JAQ10222.1 JAQ13811.1 GDIQ01083048 JAN11689.1 GBHO01045007 GBHO01045006 GBHO01045005 JAF98596.1 JAF98597.1 JAF98598.1 AGTO01000992 GDIQ01017483 JAN77254.1 GDIQ01083047 JAN11690.1 GDIQ01081546 JAN13191.1 GDIQ01091680 JAL60046.1 GDIQ01202439 JAK49286.1 GDIQ01143164 JAL08562.1 GDIQ01253978 JAJ97746.1 GDIQ01089278 JAN05459.1 GDIQ01253979 GDIQ01238096 JAK13629.1 GDIQ01249927 JAK01798.1 GDIQ01205451 JAK46274.1 GDIQ01091681 JAL60045.1 GDIQ01205450 JAK46275.1 GDIQ01249926 JAK01799.1 KZ288452 PBC25552.1 HAEB01012497 SBQ59024.1 GCES01011485 JAR74838.1 HADZ01016299 SBP80240.1 HAED01002215 SBQ88060.1 HAEI01000794 SBR73335.1 KL218030 KFP01353.1 GDIQ01079898 JAN14839.1 HADY01012657 SBP51142.1 GDIQ01203800 JAK47925.1 GDIQ01203801 JAK47924.1 KL516114 KFO88087.1 KL394174 KFP92684.1 LSYS01001520 OPJ88660.1 LMAW01003200 KQK73470.1 KK718171 KFO54764.1 HAEH01008033 SBR83412.1 HADW01010138 HADX01002250 SBP24482.1 KL215167 KFV63669.1 AYCK01000803 AYCK01000804 KL411471 KFR07765.1 KB537463 EMP33029.1 AADN05000628 KL226066 KFM07828.1 AKHW03004113 KYO31078.1 KYO31077.1 GL732535 EFX84155.1 AGCU01100386 AGCU01100387 AGCU01100388 AGCU01100389 AL833789 CU210844 KK501478 KFP15119.1 PBC25553.1 GBBI01003233 JAC15479.1
KQ460685 KPJ12793.1 JTDY01008121 KOB64727.1 JTDY01001371 KOB74045.1 KQ971363 EFA07789.1 GEZM01085937 JAV59875.1 NEVH01000597 PNF43606.1 KK852872 KDR14558.1 LJIG01009532 KRT82891.1 GEDC01001248 JAS36050.1 GEDC01023860 GEDC01021755 GEDC01018561 GEDC01014843 JAS13438.1 JAS15543.1 JAS18737.1 JAS22455.1 GL444953 EFN60365.1 AAZX01000604 GFXV01003484 MBW15289.1 GG666590 EEN51553.1 ABLF02026651 KQ434823 KZC07269.1 GL445739 EFN89140.1 DS235019 EEB10547.1 KK107570 EZA48786.1 QOIP01000005 RLU22909.1 GDHC01018461 GDHC01018295 GDHC01008407 GDHC01004818 JAQ00168.1 JAQ00334.1 JAQ10222.1 JAQ13811.1 GDIQ01083048 JAN11689.1 GBHO01045007 GBHO01045006 GBHO01045005 JAF98596.1 JAF98597.1 JAF98598.1 AGTO01000992 GDIQ01017483 JAN77254.1 GDIQ01083047 JAN11690.1 GDIQ01081546 JAN13191.1 GDIQ01091680 JAL60046.1 GDIQ01202439 JAK49286.1 GDIQ01143164 JAL08562.1 GDIQ01253978 JAJ97746.1 GDIQ01089278 JAN05459.1 GDIQ01253979 GDIQ01238096 JAK13629.1 GDIQ01249927 JAK01798.1 GDIQ01205451 JAK46274.1 GDIQ01091681 JAL60045.1 GDIQ01205450 JAK46275.1 GDIQ01249926 JAK01799.1 KZ288452 PBC25552.1 HAEB01012497 SBQ59024.1 GCES01011485 JAR74838.1 HADZ01016299 SBP80240.1 HAED01002215 SBQ88060.1 HAEI01000794 SBR73335.1 KL218030 KFP01353.1 GDIQ01079898 JAN14839.1 HADY01012657 SBP51142.1 GDIQ01203800 JAK47925.1 GDIQ01203801 JAK47924.1 KL516114 KFO88087.1 KL394174 KFP92684.1 LSYS01001520 OPJ88660.1 LMAW01003200 KQK73470.1 KK718171 KFO54764.1 HAEH01008033 SBR83412.1 HADW01010138 HADX01002250 SBP24482.1 KL215167 KFV63669.1 AYCK01000803 AYCK01000804 KL411471 KFR07765.1 KB537463 EMP33029.1 AADN05000628 KL226066 KFM07828.1 AKHW03004113 KYO31078.1 KYO31077.1 GL732535 EFX84155.1 AGCU01100386 AGCU01100387 AGCU01100388 AGCU01100389 AL833789 CU210844 KK501478 KFP15119.1 PBC25553.1 GBBI01003233 JAC15479.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000053240
UP000037510
UP000007266
+ More
UP000235965 UP000027135 UP000000311 UP000002358 UP000001554 UP000007819 UP000076420 UP000076502 UP000008237 UP000192223 UP000009046 UP000005203 UP000053097 UP000279307 UP000016665 UP000242457 UP000264820 UP000265020 UP000265100 UP000085678 UP000054308 UP000264840 UP000264800 UP000190648 UP000051836 UP000052976 UP000261420 UP000007303 UP000053875 UP000261480 UP000028760 UP000261500 UP000261360 UP000053283 UP000265120 UP000031443 UP000000539 UP000005226 UP000053286 UP000261460 UP000050525 UP000261340 UP000000305 UP000261400 UP000192220 UP000007267 UP000265040 UP000192224 UP000000437 UP000242638 UP000053119 UP000189705
UP000235965 UP000027135 UP000000311 UP000002358 UP000001554 UP000007819 UP000076420 UP000076502 UP000008237 UP000192223 UP000009046 UP000005203 UP000053097 UP000279307 UP000016665 UP000242457 UP000264820 UP000265020 UP000265100 UP000085678 UP000054308 UP000264840 UP000264800 UP000190648 UP000051836 UP000052976 UP000261420 UP000007303 UP000053875 UP000261480 UP000028760 UP000261500 UP000261360 UP000053283 UP000265120 UP000031443 UP000000539 UP000005226 UP000053286 UP000261460 UP000050525 UP000261340 UP000000305 UP000261400 UP000192220 UP000007267 UP000265040 UP000192224 UP000000437 UP000242638 UP000053119 UP000189705
Interpro
Gene 3D
ProteinModelPortal
H9JBY5
H9JBY2
A0A194PI78
A0A212FG87
A0A194R4Q1
A0A0L7KN86
+ More
A0A0L7LF60 D6WUV1 A0A1Y1KJS0 A0A2J7RRZ6 A0A067QYI6 A0A0T6B6Q9 A0A1B6EDT0 A0A1B6DA39 E2B1K9 K7IWR6 A0A2H8TMN0 C3Z6V7 J9JUE2 A0A2C9K6P6 A0A154P5R6 E2B587 A0A1W4WG18 E0VAZ1 A0A088ATM7 A0A026VZ87 A0A3L8DSQ5 A0A146KV46 A0A0P6E3M5 A0A0A9VSA9 U3JF44 A0A0P6JJ08 A0A0P6DTV0 A0A0P6D9B1 A0A0P5SL14 A0A0P5L037 A0A0P5NQP0 A0A0P5FPK1 A0A0P6CR19 A0A0P5IHA0 A0A0P5HHQ1 A0A0N8B7F1 A0A0P5S7K9 A0A0P5J486 A0A0P5G7Y3 A0A2A3E2K1 A0A1A8FLL3 A0A147A8I6 A0A3Q2Y1H6 A0A3Q2C6R7 A0A1A8CNB0 A0A1A8HUK2 A0A3P8QME6 A0A1A8NWB8 A0A3P8QKZ9 A0A1S3K5P5 A0A091I1Q1 A0A3Q2V915 A0A3Q2ZHB3 A0A0P6DIU8 A0A1A8A7P1 A0A0P5JKA5 A0A0P5KWY8 A0A091GZY1 A0A091NUU6 A0A1V4KW33 A0A0Q3SYV2 A0A091EEA5 A0A1A8PQY4 A0A1A7Y227 A0A3B4U3R2 H3CFR8 A0A093GA60 A0A3B3Y5S5 A0A096MGL3 A0A3B3W0G2 A0A3B4YEX7 A0A091VY85 A0A3P8WW56 M7B7C5 F1NIQ1 H2V487 A0A3Q2TSE6 A0A087R2X4 A0A3B4FBR3 A0A151N2U7 A0A151N2Y0 A0A3Q0SWV1 E9G951 A0A3B5AKG1 A0A2I4DBF1 K7FMX6 A0A3Q1IMD4 A0A1W5AW95 E7F7E2 A0A3P9MUN7 A0A091J299 A0A2A3E1C8 A0A1U7R9M8 A0A023F3M5
A0A0L7LF60 D6WUV1 A0A1Y1KJS0 A0A2J7RRZ6 A0A067QYI6 A0A0T6B6Q9 A0A1B6EDT0 A0A1B6DA39 E2B1K9 K7IWR6 A0A2H8TMN0 C3Z6V7 J9JUE2 A0A2C9K6P6 A0A154P5R6 E2B587 A0A1W4WG18 E0VAZ1 A0A088ATM7 A0A026VZ87 A0A3L8DSQ5 A0A146KV46 A0A0P6E3M5 A0A0A9VSA9 U3JF44 A0A0P6JJ08 A0A0P6DTV0 A0A0P6D9B1 A0A0P5SL14 A0A0P5L037 A0A0P5NQP0 A0A0P5FPK1 A0A0P6CR19 A0A0P5IHA0 A0A0P5HHQ1 A0A0N8B7F1 A0A0P5S7K9 A0A0P5J486 A0A0P5G7Y3 A0A2A3E2K1 A0A1A8FLL3 A0A147A8I6 A0A3Q2Y1H6 A0A3Q2C6R7 A0A1A8CNB0 A0A1A8HUK2 A0A3P8QME6 A0A1A8NWB8 A0A3P8QKZ9 A0A1S3K5P5 A0A091I1Q1 A0A3Q2V915 A0A3Q2ZHB3 A0A0P6DIU8 A0A1A8A7P1 A0A0P5JKA5 A0A0P5KWY8 A0A091GZY1 A0A091NUU6 A0A1V4KW33 A0A0Q3SYV2 A0A091EEA5 A0A1A8PQY4 A0A1A7Y227 A0A3B4U3R2 H3CFR8 A0A093GA60 A0A3B3Y5S5 A0A096MGL3 A0A3B3W0G2 A0A3B4YEX7 A0A091VY85 A0A3P8WW56 M7B7C5 F1NIQ1 H2V487 A0A3Q2TSE6 A0A087R2X4 A0A3B4FBR3 A0A151N2U7 A0A151N2Y0 A0A3Q0SWV1 E9G951 A0A3B5AKG1 A0A2I4DBF1 K7FMX6 A0A3Q1IMD4 A0A1W5AW95 E7F7E2 A0A3P9MUN7 A0A091J299 A0A2A3E1C8 A0A1U7R9M8 A0A023F3M5
PDB
3IRA
E-value=4.11278e-46,
Score=468
Ontologies
GO
PANTHER
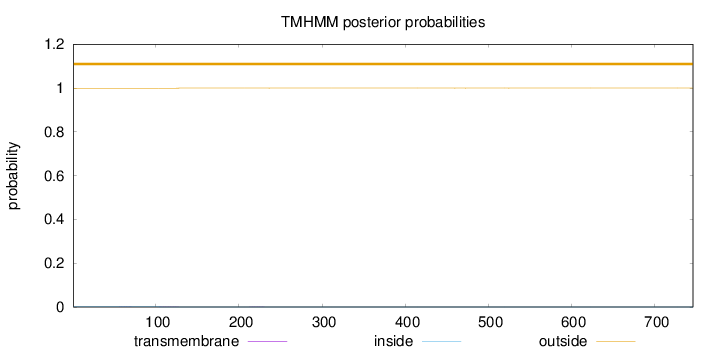
Topology
Length:
746
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01944
Exp number, first 60 AAs:
0.00096
Total prob of N-in:
0.00105
outside
1 - 746
Population Genetic Test Statistics
Pi
292.204715
Theta
170.934442
Tajima's D
-1.391418
CLR
0.807896
CSRT
0.072296385180741
Interpretation
Possibly Positive selection
