Gene
KWMTBOMO10401
Pre Gene Modal
BGIBMGA007073
Annotation
PREDICTED:_ubiquitin_carboxyl-terminal_hydrolase_nonstop_[Papilio_machaon]
Full name
Ubiquitinyl hydrolase 1
+ More
Ubiquitin carboxyl-terminal hydrolase nonstop
Ubiquitin carboxyl-terminal hydrolase nonstop
Alternative Name
Deubiquitinating enzyme nonstop
Ubiquitin thioesterase nonstop
Ubiquitin-specific-processing protease nonstop
Ubiquitin thioesterase nonstop
Ubiquitin-specific-processing protease nonstop
Location in the cell
Extracellular Reliability : 1.224 PlasmaMembrane Reliability : 1.623
Sequence
CDS
ATGAATGACAATGGTTGCATACACCTAAATAATTTTAAGGCTGCTAAAGGCACTAACCCATATAAAATCGTACATGCATTTTTTGTTTCCTGTACGTCGTATGAAGCACGGAGATACAAGGCAACATCCTGTCTCTGCTTTGCTTGTGGCCAGCCTGGACCTCGTATGCATTCATGTCTCCACTGTATCTACTTTGCATGCTATGCTGGTCATATTCAAGAACACTCCAAATCTAAGAAACACTTCCTGTATGTTGACCTTAGCTATGGAAATGTCTTCTGTGCTCAATGCCAAGACTACATTTATGATAGGGAGCTTACTGAGATCGCTGCAATACATAAGGCAAAGGAGGCTAAGTCCCTTGGTATAAGCATACCTTTTATGCCATGGTTACCAAACCATAATGAAGCAATGGCTCTTCGTCGCCTACGTAAACGGCGCCTTATCAGTCCTCATACTACAATAGGTTTACGGGGCTTACAGAACCTAGGATCAACATGCTTCATGAACTGCATTGTTCAGACACTAATTCACACTCCACTCCTGAGAGATTATTTCTTAGCAGAAAAACACAAATGTAAGTCGCAAAGTTCTAGCAAATGCTTGGTGTGTGAAGTTTCCAAACTATTTCAGGAGTTTTACTCTGGAGCAAAAACACCGCTGACTTTGCATCGTCTATTACACTTAATATGGACTCACGCCCGTCACTTGGCTGGATATGAACAACAGGATGCTCATGAATTCTTCATTGCCACTCTTGATGTGCTGCATAGACATTGCATGAATGGAGTAGAAGATACAGAAAAAAAGGAAAATGGGCGTTGTAATTGCATTATCGATCAGATATTCACCGGCGGATTACAAAGCGACGTCGTGTGCACCTCCTGCTCTGGAGTCTCCACTACCATCGATCCGTTCTGGGACATCAGTCTTGACGTGGCCGGGCCGGGTTCCCTGCAAGCTTGCTTGGAACGTTTCACCAGAGCCGAACACCTGGGCTCGGCGGCGAAAATAAAATGCTCCAACTGTCGCGCCTACCGTGAATCCACCAAGCAGCTGACTTTAGAGACTTTGCCGATCGTAGCGAGCTTCCACCTCAAACGTTTCGAGCATTCGTCTCAGATCGATCGGAAGATATCCGCCTTCGTATCGTTCCCGGCTGAGTTAGACATGACTCCGTTTATGTCGTCGCATCGTCGTGCCGTCGACGTGGGCGCGGCCGAGAACAATAACACCCCGGAAGGAATGTTCGAGGACAACCGCTACTCTCTGTTCGCGGTGGTCAATCACTTGGGTTCGCTGGACGCAGGGCACTACACCGCATACGTCAGGCAAATGAAAGGAAGCTGGTTCAAATGCGACGACCACATGATTACTAGGGCGTCACTTCGAGAGGTTTTAGATAGCGAAGGATATCTTCTGTTTTATCACAAGACTGTGTTGGAGTATGAATGCGACGTTTCTTAA
Protein
MNDNGCIHLNNFKAAKGTNPYKIVHAFFVSCTSYEARRYKATSCLCFACGQPGPRMHSCLHCIYFACYAGHIQEHSKSKKHFLYVDLSYGNVFCAQCQDYIYDRELTEIAAIHKAKEAKSLGISIPFMPWLPNHNEAMALRRLRKRRLISPHTTIGLRGLQNLGSTCFMNCIVQTLIHTPLLRDYFLAEKHKCKSQSSSKCLVCEVSKLFQEFYSGAKTPLTLHRLLHLIWTHARHLAGYEQQDAHEFFIATLDVLHRHCMNGVEDTEKKENGRCNCIIDQIFTGGLQSDVVCTSCSGVSTTIDPFWDISLDVAGPGSLQACLERFTRAEHLGSAAKIKCSNCRAYRESTKQLTLETLPIVASFHLKRFEHSSQIDRKISAFVSFPAELDMTPFMSSHRRAVDVGAAENNNTPEGMFEDNRYSLFAVVNHLGSLDAGHYTAYVRQMKGSWFKCDDHMITRASLREVLDSEGYLLFYHKTVLEYECDVS
Summary
Description
Histone deubiquitinating component of the transcription regulatory histone acetylation (HAT) complex SAGA. Catalyzes the deubiquitination of histone H2B, thereby acting as a coactivator in a large subset of genes. Required to counteract heterochromatin silencing. Controls the development of neuronal connectivity in visual system by being required for accurate axon targeting in the optic lobe. Required for expression of ecdysone-induced genes such as br/broad.
Catalytic Activity
Thiol-dependent hydrolysis of ester, thioester, amide, peptide and isopeptide bonds formed by the C-terminal Gly of ubiquitin (a 76-residue protein attached to proteins as an intracellular targeting signal).
Subunit
Component of the SAGA transcription coactivator-HAT complex, at least composed of Ada2b, not/nonstop, Pcaf/Gcn5, Sgf11 and Spt3.
Similarity
Belongs to the peptidase C19 family.
Belongs to the peptidase C19 family. UBP8 subfamily.
Belongs to the peptidase C19 family. UBP8 subfamily.
Keywords
Activator
Chromatin regulator
Complete proteome
Hydrolase
Metal-binding
Nucleus
Protease
Reference proteome
Thiol protease
Transcription
Transcription regulation
Ubl conjugation pathway
Zinc
Zinc-finger
Feature
chain Ubiquitinyl hydrolase 1
Uniprot
A0A3S2LD34
A0A1E1WUC5
A0A194R534
A0A212FG94
A0A194PHX1
A0A2H1V7A7
+ More
H9JC28 A0A2A4IT90 A0A0L7REW9 A0A154P1K6 E2BHH1 A0A2A3EBC8 A0A088AMI7 A0A0C9QS82 A0A0N0U567 E9FWK2 A0A1B6KA10 A0A1B6JZA4 A0A0N8CFR9 A0A026X152 E2APY7 A0A0N8ASS7 A0A0P4YBV7 K7IYF3 A0A195DUN1 F4W884 A0A151HYV6 A0A158NSQ6 A0A151X0D1 A0A195CB19 A0A0N8D386 A0A0P4X4U1 A0A164VLS5 A0A0P5AIY8 A0A195EWI6 A0A1I8M5Q4 A0A1A9ZMV3 A0A1B0BKB3 A0A1I8M5Q9 A0A0L0CGW0 A0A1I8NM89 A0A1A9W3D5 A0A1A9Y9T6 A0A1B6C3K7 V5IEC6 B4L0D4 A0A182GN74 B3M888 B4LID4 A0A1W4W4G0 A0A293LWW2 A0A0J9UNT1 B3NHQ9 A0A3B0JAI2 Q2LYY5 B4N443 Q9VVR1 A0A1B0CL75 B4QPU1 B4PJ09 V4B961 A0A1E1XFK5 A0A224ZB43 A0A1E1XSM8 A0A131Z031 A0A0P5E6G4 A0A3P9B216 A0A3P9HMA2 A0A2R2MQJ5 A0A0N8AND0 M4AS42 A0A1B0F9K9 A0A1A9VMN7 A0A3B4FDH4 A0A3Q1GAW8 A0A3Q3C6N4 A0A3Q4HQ24 A0A3B3V445 A0A3P8QP70 K9IL10 A0A3B3XQC2 A0A087YJ29 A0A3Q1BMH9 H3CPM5 A0A3P8TSC6 A0A286ZK36 A0A2K6F1P9 A0A1A8FNJ7 A0A3P9QD20 A0A2K6F1Q1 H2V462 A0A1A8MCS6 A0A1A8RD98 A0A1A8KRA3 A0A1A8CYS4 A0A1A8U6K3 H3A8J7 A0A0A1XSX4 F7BCG1 M3W0I5
H9JC28 A0A2A4IT90 A0A0L7REW9 A0A154P1K6 E2BHH1 A0A2A3EBC8 A0A088AMI7 A0A0C9QS82 A0A0N0U567 E9FWK2 A0A1B6KA10 A0A1B6JZA4 A0A0N8CFR9 A0A026X152 E2APY7 A0A0N8ASS7 A0A0P4YBV7 K7IYF3 A0A195DUN1 F4W884 A0A151HYV6 A0A158NSQ6 A0A151X0D1 A0A195CB19 A0A0N8D386 A0A0P4X4U1 A0A164VLS5 A0A0P5AIY8 A0A195EWI6 A0A1I8M5Q4 A0A1A9ZMV3 A0A1B0BKB3 A0A1I8M5Q9 A0A0L0CGW0 A0A1I8NM89 A0A1A9W3D5 A0A1A9Y9T6 A0A1B6C3K7 V5IEC6 B4L0D4 A0A182GN74 B3M888 B4LID4 A0A1W4W4G0 A0A293LWW2 A0A0J9UNT1 B3NHQ9 A0A3B0JAI2 Q2LYY5 B4N443 Q9VVR1 A0A1B0CL75 B4QPU1 B4PJ09 V4B961 A0A1E1XFK5 A0A224ZB43 A0A1E1XSM8 A0A131Z031 A0A0P5E6G4 A0A3P9B216 A0A3P9HMA2 A0A2R2MQJ5 A0A0N8AND0 M4AS42 A0A1B0F9K9 A0A1A9VMN7 A0A3B4FDH4 A0A3Q1GAW8 A0A3Q3C6N4 A0A3Q4HQ24 A0A3B3V445 A0A3P8QP70 K9IL10 A0A3B3XQC2 A0A087YJ29 A0A3Q1BMH9 H3CPM5 A0A3P8TSC6 A0A286ZK36 A0A2K6F1P9 A0A1A8FNJ7 A0A3P9QD20 A0A2K6F1Q1 H2V462 A0A1A8MCS6 A0A1A8RD98 A0A1A8KRA3 A0A1A8CYS4 A0A1A8U6K3 H3A8J7 A0A0A1XSX4 F7BCG1 M3W0I5
EC Number
3.4.19.12
Pubmed
26354079
22118469
19121390
20798317
21292972
24508170
+ More
30249741 20075255 21719571 21347285 25315136 26108605 25765539 17994087 26483478 22936249 15632085 11182084 10731132 12537572 12537569 18188155 18206972 17550304 23254933 28503490 28797301 29209593 26830274 25186727 17554307 26383154 23542700 15496914 21551351 9215903 25830018 17495919 17975172
30249741 20075255 21719571 21347285 25315136 26108605 25765539 17994087 26483478 22936249 15632085 11182084 10731132 12537572 12537569 18188155 18206972 17550304 23254933 28503490 28797301 29209593 26830274 25186727 17554307 26383154 23542700 15496914 21551351 9215903 25830018 17495919 17975172
EMBL
RSAL01000218
RVE44093.1
GDQN01000663
JAT90391.1
KQ460685
KPJ12792.1
+ More
AGBW02008703 OWR52747.1 KQ459603 KPI93016.1 ODYU01000807 SOQ36144.1 BABH01014789 BABH01014790 NWSH01008640 PCG62464.1 KQ414608 KOC69375.1 KQ434796 KZC05723.1 GL448301 EFN84824.1 KZ288313 PBC28361.1 GBYB01003512 JAG73279.1 KQ435791 KOX74394.1 GL732526 EFX88419.1 GEBQ01031688 GEBQ01029188 JAT08289.1 JAT10789.1 GECU01003177 JAT04530.1 GDIP01136095 GDIQ01085601 JAL67619.1 JAN09136.1 KK107046 QOIP01000004 EZA61731.1 RLU24206.1 GL441643 EFN64504.1 GDIQ01246443 JAK05282.1 GDIP01233339 JAI90062.1 KQ980322 KYN16583.1 GL887898 EGI69579.1 KQ976723 KYM76672.1 ADTU01025108 KQ982624 KYQ53690.1 KQ978023 KYM98002.1 GDIQ01092455 GDIP01073159 JAL59271.1 JAM30556.1 GDIP01246356 JAI77045.1 LRGB01001361 KZS12448.1 GDIP01198344 JAJ25058.1 KQ981948 KYN32640.1 JXJN01015869 JRES01000413 KNC31476.1 GEDC01029324 JAS07974.1 GANP01010422 JAB74046.1 CH933809 EDW19103.2 JXUM01014567 KQ560407 KXJ82566.1 CH902618 EDV41027.2 CH940647 EDW70721.2 GFWV01007709 MAA32439.1 CM002912 KMZ00416.1 CH954178 EDV51854.1 OUUW01000002 SPP77002.1 CH379069 EAL29724.3 CH964095 EDW78917.2 AF179590 AE014296 AY058707 AJWK01017007 AJWK01017008 AJWK01017009 AJWK01017010 CM000363 EDX10969.1 CM000159 EDW94600.2 KB199905 ESP03856.1 GFAC01001166 JAT98022.1 GFPF01013058 MAA24204.1 GFAA01001112 JAU02323.1 GEDV01003764 JAP84793.1 GDIP01145900 JAJ77502.1 GDIQ01258819 JAJ92905.1 CCAG010015211 GABZ01005510 JAA48015.1 AYCK01000799 AEMK02000083 HAEB01013906 SBQ60433.1 HAEF01012985 SBR54144.1 HAEH01015641 HAEI01014619 SBS03249.1 HAED01006291 HAEE01014867 SBR34917.1 HADZ01021021 HAEA01000576 SBP84962.1 HADY01022142 HAEJ01003487 SBS43944.1 AFYH01177354 AFYH01177355 AFYH01177356 AFYH01177357 AFYH01177358 AFYH01177359 AFYH01177360 AFYH01177361 AFYH01177362 AFYH01177363 GBXI01000277 JAD14015.1 AANG04000740
AGBW02008703 OWR52747.1 KQ459603 KPI93016.1 ODYU01000807 SOQ36144.1 BABH01014789 BABH01014790 NWSH01008640 PCG62464.1 KQ414608 KOC69375.1 KQ434796 KZC05723.1 GL448301 EFN84824.1 KZ288313 PBC28361.1 GBYB01003512 JAG73279.1 KQ435791 KOX74394.1 GL732526 EFX88419.1 GEBQ01031688 GEBQ01029188 JAT08289.1 JAT10789.1 GECU01003177 JAT04530.1 GDIP01136095 GDIQ01085601 JAL67619.1 JAN09136.1 KK107046 QOIP01000004 EZA61731.1 RLU24206.1 GL441643 EFN64504.1 GDIQ01246443 JAK05282.1 GDIP01233339 JAI90062.1 KQ980322 KYN16583.1 GL887898 EGI69579.1 KQ976723 KYM76672.1 ADTU01025108 KQ982624 KYQ53690.1 KQ978023 KYM98002.1 GDIQ01092455 GDIP01073159 JAL59271.1 JAM30556.1 GDIP01246356 JAI77045.1 LRGB01001361 KZS12448.1 GDIP01198344 JAJ25058.1 KQ981948 KYN32640.1 JXJN01015869 JRES01000413 KNC31476.1 GEDC01029324 JAS07974.1 GANP01010422 JAB74046.1 CH933809 EDW19103.2 JXUM01014567 KQ560407 KXJ82566.1 CH902618 EDV41027.2 CH940647 EDW70721.2 GFWV01007709 MAA32439.1 CM002912 KMZ00416.1 CH954178 EDV51854.1 OUUW01000002 SPP77002.1 CH379069 EAL29724.3 CH964095 EDW78917.2 AF179590 AE014296 AY058707 AJWK01017007 AJWK01017008 AJWK01017009 AJWK01017010 CM000363 EDX10969.1 CM000159 EDW94600.2 KB199905 ESP03856.1 GFAC01001166 JAT98022.1 GFPF01013058 MAA24204.1 GFAA01001112 JAU02323.1 GEDV01003764 JAP84793.1 GDIP01145900 JAJ77502.1 GDIQ01258819 JAJ92905.1 CCAG010015211 GABZ01005510 JAA48015.1 AYCK01000799 AEMK02000083 HAEB01013906 SBQ60433.1 HAEF01012985 SBR54144.1 HAEH01015641 HAEI01014619 SBS03249.1 HAED01006291 HAEE01014867 SBR34917.1 HADZ01021021 HAEA01000576 SBP84962.1 HADY01022142 HAEJ01003487 SBS43944.1 AFYH01177354 AFYH01177355 AFYH01177356 AFYH01177357 AFYH01177358 AFYH01177359 AFYH01177360 AFYH01177361 AFYH01177362 AFYH01177363 GBXI01000277 JAD14015.1 AANG04000740
Proteomes
UP000283053
UP000053240
UP000007151
UP000053268
UP000005204
UP000218220
+ More
UP000053825 UP000076502 UP000008237 UP000242457 UP000005203 UP000053105 UP000000305 UP000053097 UP000279307 UP000000311 UP000002358 UP000078492 UP000007755 UP000078540 UP000005205 UP000075809 UP000078542 UP000076858 UP000078541 UP000095301 UP000092445 UP000092460 UP000037069 UP000095300 UP000091820 UP000092443 UP000009192 UP000069940 UP000249989 UP000007801 UP000008792 UP000192221 UP000008711 UP000268350 UP000001819 UP000007798 UP000000803 UP000092461 UP000000304 UP000002282 UP000030746 UP000265160 UP000265200 UP000085678 UP000002852 UP000092444 UP000078200 UP000261460 UP000257200 UP000264840 UP000261580 UP000261500 UP000265100 UP000261480 UP000028760 UP000257160 UP000007303 UP000265080 UP000008227 UP000233160 UP000242638 UP000005226 UP000008672 UP000002280 UP000011712
UP000053825 UP000076502 UP000008237 UP000242457 UP000005203 UP000053105 UP000000305 UP000053097 UP000279307 UP000000311 UP000002358 UP000078492 UP000007755 UP000078540 UP000005205 UP000075809 UP000078542 UP000076858 UP000078541 UP000095301 UP000092445 UP000092460 UP000037069 UP000095300 UP000091820 UP000092443 UP000009192 UP000069940 UP000249989 UP000007801 UP000008792 UP000192221 UP000008711 UP000268350 UP000001819 UP000007798 UP000000803 UP000092461 UP000000304 UP000002282 UP000030746 UP000265160 UP000265200 UP000085678 UP000002852 UP000092444 UP000078200 UP000261460 UP000257200 UP000264840 UP000261580 UP000261500 UP000265100 UP000261480 UP000028760 UP000257160 UP000007303 UP000265080 UP000008227 UP000233160 UP000242638 UP000005226 UP000008672 UP000002280 UP000011712
Interpro
Gene 3D
ProteinModelPortal
A0A3S2LD34
A0A1E1WUC5
A0A194R534
A0A212FG94
A0A194PHX1
A0A2H1V7A7
+ More
H9JC28 A0A2A4IT90 A0A0L7REW9 A0A154P1K6 E2BHH1 A0A2A3EBC8 A0A088AMI7 A0A0C9QS82 A0A0N0U567 E9FWK2 A0A1B6KA10 A0A1B6JZA4 A0A0N8CFR9 A0A026X152 E2APY7 A0A0N8ASS7 A0A0P4YBV7 K7IYF3 A0A195DUN1 F4W884 A0A151HYV6 A0A158NSQ6 A0A151X0D1 A0A195CB19 A0A0N8D386 A0A0P4X4U1 A0A164VLS5 A0A0P5AIY8 A0A195EWI6 A0A1I8M5Q4 A0A1A9ZMV3 A0A1B0BKB3 A0A1I8M5Q9 A0A0L0CGW0 A0A1I8NM89 A0A1A9W3D5 A0A1A9Y9T6 A0A1B6C3K7 V5IEC6 B4L0D4 A0A182GN74 B3M888 B4LID4 A0A1W4W4G0 A0A293LWW2 A0A0J9UNT1 B3NHQ9 A0A3B0JAI2 Q2LYY5 B4N443 Q9VVR1 A0A1B0CL75 B4QPU1 B4PJ09 V4B961 A0A1E1XFK5 A0A224ZB43 A0A1E1XSM8 A0A131Z031 A0A0P5E6G4 A0A3P9B216 A0A3P9HMA2 A0A2R2MQJ5 A0A0N8AND0 M4AS42 A0A1B0F9K9 A0A1A9VMN7 A0A3B4FDH4 A0A3Q1GAW8 A0A3Q3C6N4 A0A3Q4HQ24 A0A3B3V445 A0A3P8QP70 K9IL10 A0A3B3XQC2 A0A087YJ29 A0A3Q1BMH9 H3CPM5 A0A3P8TSC6 A0A286ZK36 A0A2K6F1P9 A0A1A8FNJ7 A0A3P9QD20 A0A2K6F1Q1 H2V462 A0A1A8MCS6 A0A1A8RD98 A0A1A8KRA3 A0A1A8CYS4 A0A1A8U6K3 H3A8J7 A0A0A1XSX4 F7BCG1 M3W0I5
H9JC28 A0A2A4IT90 A0A0L7REW9 A0A154P1K6 E2BHH1 A0A2A3EBC8 A0A088AMI7 A0A0C9QS82 A0A0N0U567 E9FWK2 A0A1B6KA10 A0A1B6JZA4 A0A0N8CFR9 A0A026X152 E2APY7 A0A0N8ASS7 A0A0P4YBV7 K7IYF3 A0A195DUN1 F4W884 A0A151HYV6 A0A158NSQ6 A0A151X0D1 A0A195CB19 A0A0N8D386 A0A0P4X4U1 A0A164VLS5 A0A0P5AIY8 A0A195EWI6 A0A1I8M5Q4 A0A1A9ZMV3 A0A1B0BKB3 A0A1I8M5Q9 A0A0L0CGW0 A0A1I8NM89 A0A1A9W3D5 A0A1A9Y9T6 A0A1B6C3K7 V5IEC6 B4L0D4 A0A182GN74 B3M888 B4LID4 A0A1W4W4G0 A0A293LWW2 A0A0J9UNT1 B3NHQ9 A0A3B0JAI2 Q2LYY5 B4N443 Q9VVR1 A0A1B0CL75 B4QPU1 B4PJ09 V4B961 A0A1E1XFK5 A0A224ZB43 A0A1E1XSM8 A0A131Z031 A0A0P5E6G4 A0A3P9B216 A0A3P9HMA2 A0A2R2MQJ5 A0A0N8AND0 M4AS42 A0A1B0F9K9 A0A1A9VMN7 A0A3B4FDH4 A0A3Q1GAW8 A0A3Q3C6N4 A0A3Q4HQ24 A0A3B3V445 A0A3P8QP70 K9IL10 A0A3B3XQC2 A0A087YJ29 A0A3Q1BMH9 H3CPM5 A0A3P8TSC6 A0A286ZK36 A0A2K6F1P9 A0A1A8FNJ7 A0A3P9QD20 A0A2K6F1Q1 H2V462 A0A1A8MCS6 A0A1A8RD98 A0A1A8KRA3 A0A1A8CYS4 A0A1A8U6K3 H3A8J7 A0A0A1XSX4 F7BCG1 M3W0I5
PDB
3M99
E-value=1.31635e-51,
Score=514
Ontologies
GO
GO:0046872
GO:0036459
GO:0006511
GO:0008270
GO:0016579
GO:0016021
GO:0004843
GO:0016578
GO:0045893
GO:0008347
GO:0021782
GO:0007411
GO:0000124
GO:0071819
GO:0007412
GO:0005634
GO:0006355
GO:0006325
GO:0003676
GO:0031047
GO:0016607
GO:0019899
GO:0010485
GO:0016574
GO:0045931
GO:0030374
GO:0004674
GO:0006415
GO:0006418
GO:0003723
GO:0003824
Topology
Subcellular location
Nucleus
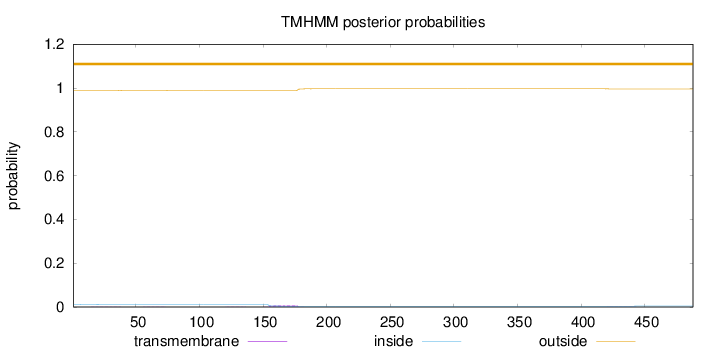
Length:
488
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.22044
Exp number, first 60 AAs:
0.01273
Total prob of N-in:
0.01225
outside
1 - 488
Population Genetic Test Statistics
Pi
187.350924
Theta
154.195552
Tajima's D
0.920101
CLR
0
CSRT
0.63916804159792
Interpretation
Uncertain
