Gene
KWMTBOMO10398 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007072
Annotation
yellow-b_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.463 PlasmaMembrane Reliability : 1.067
Sequence
CDS
ATGCGCTACACGCTGACATCGCTCTCGATGATGATAGCTATAGTGCTCCTCGCCGCCGTCGCCGCCGCGGCCGCGCAAAACAAGACAACACTCCCGAAGACGGATGTTAACCCCCGACAGTTTCGCGTTGTCTACGAATGGAACGCAATCGATTTCGAATGGACGTCTCCAGAAGATCGCGAGGCCTACCTCAATACGAGCCAATATATACCCCAAAACGTCCTAATATCTGGCATAAACTTCTACGGCGAAAACTTGTTCCTGACTATGCCGAGGATGTTAGCCGGAGTGCCGGCGACGTTAGCGACAATACCGATACAGCAAGTAAACACGGCGCCGAAACTGAAGCCTTTCCCGAGTTGGGCCGATAACGCCATCGGTAACTGCAACGCTCTACAATTCGTTCAAAACATAGAAATTGATAGGAACGGTATTATGTGGATTCTCGATAACGGACGAGTGGGCACTCTTACCCAGAACCCTGACCCGAAGTGTCCTCCGTCAATCGTGCTCATCGATCTTAAATCTGAAAAACTTGAAATGGAACGGATTCCGTTCCCGCCAGAAACGGTTAATCCCAACACGACTTACCTCAATGATCTCGTCGTAGACAATCGAGACGGTGATTACGCGTACATAACCGATAACAGTGCCGTGGATCCAGGTATAATTGTCTTCCGCAGAAGTGACAAGAGATCGTGGAAACTCCGTGACGGAAAATCGATGAGGGCCGCAAATGACGCTACGTTCTTCCGCATAAACGGCACAACGGTCAATCTTCCGGTTAACATCGACGGAATAGCCTTAGGACCGCAATATGTGACGGAAGACGGTAAAGTAGACAGAAAAATATATTATAGTCCTCTTGCTAGTTTTCATCTGTACGCCATAAACGCGTCAGTTTTACAAAACGAATCGCTCGCACCATGGGGCGATGGAGCTTTACAGCCCTACGTCATTGATCTTGGCACTAAAGCTTCTCAAACAGATGGCATGAAAATGGATTCAGCGGGCAACTTATACTACGGCCTTTTAGGTATTGCATCGATCGCTGAATGGAATTCAACGCTAGATTTTCACGTCGGCCAAAGAACGATTGCACGCGATTCTAACTACATACAATGGGTGGACCGTTTCACTTTTGACGACAAAGGAAACGTTTACGTCATAATCAACAGGCTGTACAACTTCGTGAAAAATCAGTTATCTCTGGACGAAGTTAACTACAGAATTTTGCGATCGCACACGGGATCGAAAAGCTACGTATTCGGGGAAGACGTGACGCCGGTAATGAAGCCGGCTTACGACGCAGCAACGGTTCCAACCCTGGGAGCGTCGCTGTTGGCACTTGCGCTAGCACACCTGCTTGCTTAG
Protein
MRYTLTSLSMMIAIVLLAAVAAAAAQNKTTLPKTDVNPRQFRVVYEWNAIDFEWTSPEDREAYLNTSQYIPQNVLISGINFYGENLFLTMPRMLAGVPATLATIPIQQVNTAPKLKPFPSWADNAIGNCNALQFVQNIEIDRNGIMWILDNGRVGTLTQNPDPKCPPSIVLIDLKSEKLEMERIPFPPETVNPNTTYLNDLVVDNRDGDYAYITDNSAVDPGIIVFRRSDKRSWKLRDGKSMRAANDATFFRINGTTVNLPVNIDGIALGPQYVTEDGKVDRKIYYSPLASFHLYAINASVLQNESLAPWGDGALQPYVIDLGTKASQTDGMKMDSAGNLYYGLLGIASIAEWNSTLDFHVGQRTIARDSNYIQWVDRFTFDDKGNVYVIINRLYNFVKNQLSLDEVNYRILRSHTGSKSYVFGEDVTPVMKPAYDAATVPTLGASLLALALAHLLA
Summary
Uniprot
H9B419
Q2HZG2
H9JC27
A0A2H1WVM2
I4DJT6
A0A194R5J9
+ More
A0A0L7KV09 A0A212FG92 S4NXS5 F1AAP3 E3SWU4 E3SWV1 Q7Q8V5 A0A182U9C3 A0A182WUS9 A0A182LQ05 A0A182HFT7 A0A182VIE3 A0A182SY88 A0A1S4H4B4 A0A182K9W5 A0A1Q3FZM9 A0A1Q3FZL8 A0A1Q3FZS1 A0A1Q3FZQ3 A0A1Q3FZI2 A0A182PJK2 A0A1Q3FZI1 A0A182M3Y0 A0A182YF62 A0A1L8DNH0 A0A182VT89 A0A1L8DNC9 A0A182JE23 A0A182QKC9 N6UB80 A0A182RZ12 A0A182MYB3 A0A023EUE1 A0A182H3N7 A0A023EUI4 A0A1J1ISZ4 A0A023ETP7 A0A023EW07 A0A2M4BJ17 A0A2M4BJ01 W5JDJ6 A0A2M4ATQ7 A0A2M4BMB2 A0A2M4BLF6 A0A182FUI0 A0A290GAZ2 A0A067RKF2 A0A1Y1LM04 A0A2A3EAP0 V9IFQ1 A0A088AMI6 A0A0N0BG79 A0A2J7PLV4 D6WYJ5 D1LZL0 A0A026X0A9 E2BHH2 A0A154P303 A0A195CRN3 A0A310S9B6 E9J0G9 A0A151WRD6 E2ABX0 A0A151I4U0 A0A1W4XS25 A0A084WAU3 A0A0C9QZK4 E0VCA3 A0A0L7RF83 A0A232F2M5 A0A1B0DC19 K7IYF4 K7J772 A0A232EUG6 K7IYA1 A0A232F6B1 A0A1D2MCF9 U4TX99 N6TS19 A0A1W4XEE6 A0A290GC51 D1LZL2 D6WH81 A0A1W4X3L4 K7J7C1 A0A232EVL6 D6WH83 A0A290GSI7 D6WH82
A0A0L7KV09 A0A212FG92 S4NXS5 F1AAP3 E3SWU4 E3SWV1 Q7Q8V5 A0A182U9C3 A0A182WUS9 A0A182LQ05 A0A182HFT7 A0A182VIE3 A0A182SY88 A0A1S4H4B4 A0A182K9W5 A0A1Q3FZM9 A0A1Q3FZL8 A0A1Q3FZS1 A0A1Q3FZQ3 A0A1Q3FZI2 A0A182PJK2 A0A1Q3FZI1 A0A182M3Y0 A0A182YF62 A0A1L8DNH0 A0A182VT89 A0A1L8DNC9 A0A182JE23 A0A182QKC9 N6UB80 A0A182RZ12 A0A182MYB3 A0A023EUE1 A0A182H3N7 A0A023EUI4 A0A1J1ISZ4 A0A023ETP7 A0A023EW07 A0A2M4BJ17 A0A2M4BJ01 W5JDJ6 A0A2M4ATQ7 A0A2M4BMB2 A0A2M4BLF6 A0A182FUI0 A0A290GAZ2 A0A067RKF2 A0A1Y1LM04 A0A2A3EAP0 V9IFQ1 A0A088AMI6 A0A0N0BG79 A0A2J7PLV4 D6WYJ5 D1LZL0 A0A026X0A9 E2BHH2 A0A154P303 A0A195CRN3 A0A310S9B6 E9J0G9 A0A151WRD6 E2ABX0 A0A151I4U0 A0A1W4XS25 A0A084WAU3 A0A0C9QZK4 E0VCA3 A0A0L7RF83 A0A232F2M5 A0A1B0DC19 K7IYF4 K7J772 A0A232EUG6 K7IYA1 A0A232F6B1 A0A1D2MCF9 U4TX99 N6TS19 A0A1W4XEE6 A0A290GC51 D1LZL2 D6WH81 A0A1W4X3L4 K7J7C1 A0A232EVL6 D6WH83 A0A290GSI7 D6WH82
Pubmed
EMBL
JN977504
AFC87784.1
DQ358083
ABC96698.1
BABH01014783
ODYU01011354
+ More
SOQ57047.1 AK401554 KQ459603 BAM18176.1 KPI93012.1 KQ460685 KPJ12789.1 JTDY01005585 KOB66859.1 AGBW02008703 OWR52749.1 GAIX01014070 JAA78490.1 GU063844 ADX87362.1 GU128941 ADD60457.1 GU128948 ADD60458.1 AAAB01008933 EAA09869.4 APCN01005579 GFDL01002065 JAV32980.1 GFDL01002041 JAV33004.1 GFDL01002043 JAV33002.1 GFDL01002063 JAV32982.1 GFDL01002066 JAV32979.1 GFDL01002064 JAV32981.1 AXCM01002855 GFDF01006123 JAV07961.1 GFDF01006122 JAV07962.1 AXCN02002049 APGK01035666 APGK01035667 APGK01035668 APGK01035669 KB740928 ENN77911.1 GAPW01001062 JAC12536.1 JXUM01107912 JXUM01107913 JXUM01107914 JXUM01107915 KQ565183 KXJ71245.1 GAPW01000981 JAC12617.1 CVRI01000059 CRL03363.1 GAPW01000878 JAC12720.1 GAPW01001084 JAC12514.1 GGFJ01003904 MBW53045.1 GGFJ01003905 MBW53046.1 ADMH02001479 ETN62427.1 GGFK01010850 MBW44171.1 GGFJ01004767 MBW53908.1 GGFJ01004768 MBW53909.1 KY221851 ATB56349.1 KK852421 KDR24297.1 GEZM01051777 JAV74704.1 KZ288313 PBC28362.1 JR041710 AEY59497.1 KQ435791 KOX74392.1 NEVH01024423 PNF17318.1 KQ971362 EFA07870.1 GU111771 ACY71064.1 KK107046 QOIP01000004 EZA61730.1 RLU24200.1 GL448301 EFN84825.1 KQ434796 KZC05724.1 KQ977349 KYN03366.1 KQ767621 OAD53378.1 GL767474 EFZ13680.1 KQ982807 KYQ50367.1 GL438386 EFN69065.1 KQ976460 KYM84753.1 ATLV01022266 ATLV01022267 KE525331 KFB47337.1 GBYB01000061 JAG69828.1 DS235051 EEB11025.1 KQ414608 KOC69376.1 NNAY01001206 OXU24740.1 AJVK01030520 NNAY01002129 OXU22003.1 NNAY01000875 OXU26112.1 LJIJ01001848 ODM90687.1 KB631694 ERL85417.1 APGK01056595 APGK01056596 APGK01056597 KB741277 ENN71076.1 KY221859 ATB56357.1 GU111773 ACY71066.1 KQ971330 EFA00985.1 NNAY01001977 OXU22396.1 EEZ99739.1 KY221848 ATB56346.1 EEZ99740.1
SOQ57047.1 AK401554 KQ459603 BAM18176.1 KPI93012.1 KQ460685 KPJ12789.1 JTDY01005585 KOB66859.1 AGBW02008703 OWR52749.1 GAIX01014070 JAA78490.1 GU063844 ADX87362.1 GU128941 ADD60457.1 GU128948 ADD60458.1 AAAB01008933 EAA09869.4 APCN01005579 GFDL01002065 JAV32980.1 GFDL01002041 JAV33004.1 GFDL01002043 JAV33002.1 GFDL01002063 JAV32982.1 GFDL01002066 JAV32979.1 GFDL01002064 JAV32981.1 AXCM01002855 GFDF01006123 JAV07961.1 GFDF01006122 JAV07962.1 AXCN02002049 APGK01035666 APGK01035667 APGK01035668 APGK01035669 KB740928 ENN77911.1 GAPW01001062 JAC12536.1 JXUM01107912 JXUM01107913 JXUM01107914 JXUM01107915 KQ565183 KXJ71245.1 GAPW01000981 JAC12617.1 CVRI01000059 CRL03363.1 GAPW01000878 JAC12720.1 GAPW01001084 JAC12514.1 GGFJ01003904 MBW53045.1 GGFJ01003905 MBW53046.1 ADMH02001479 ETN62427.1 GGFK01010850 MBW44171.1 GGFJ01004767 MBW53908.1 GGFJ01004768 MBW53909.1 KY221851 ATB56349.1 KK852421 KDR24297.1 GEZM01051777 JAV74704.1 KZ288313 PBC28362.1 JR041710 AEY59497.1 KQ435791 KOX74392.1 NEVH01024423 PNF17318.1 KQ971362 EFA07870.1 GU111771 ACY71064.1 KK107046 QOIP01000004 EZA61730.1 RLU24200.1 GL448301 EFN84825.1 KQ434796 KZC05724.1 KQ977349 KYN03366.1 KQ767621 OAD53378.1 GL767474 EFZ13680.1 KQ982807 KYQ50367.1 GL438386 EFN69065.1 KQ976460 KYM84753.1 ATLV01022266 ATLV01022267 KE525331 KFB47337.1 GBYB01000061 JAG69828.1 DS235051 EEB11025.1 KQ414608 KOC69376.1 NNAY01001206 OXU24740.1 AJVK01030520 NNAY01002129 OXU22003.1 NNAY01000875 OXU26112.1 LJIJ01001848 ODM90687.1 KB631694 ERL85417.1 APGK01056595 APGK01056596 APGK01056597 KB741277 ENN71076.1 KY221859 ATB56357.1 GU111773 ACY71066.1 KQ971330 EFA00985.1 NNAY01001977 OXU22396.1 EEZ99739.1 KY221848 ATB56346.1 EEZ99740.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000037510
UP000007151
UP000007062
+ More
UP000075902 UP000076407 UP000075882 UP000075840 UP000075903 UP000075901 UP000075881 UP000075885 UP000075883 UP000076408 UP000075920 UP000075880 UP000075886 UP000019118 UP000075900 UP000075884 UP000069940 UP000249989 UP000183832 UP000000673 UP000069272 UP000027135 UP000242457 UP000005203 UP000053105 UP000235965 UP000007266 UP000053097 UP000279307 UP000008237 UP000076502 UP000078542 UP000075809 UP000000311 UP000078540 UP000192223 UP000030765 UP000009046 UP000053825 UP000215335 UP000092462 UP000002358 UP000094527 UP000030742
UP000075902 UP000076407 UP000075882 UP000075840 UP000075903 UP000075901 UP000075881 UP000075885 UP000075883 UP000076408 UP000075920 UP000075880 UP000075886 UP000019118 UP000075900 UP000075884 UP000069940 UP000249989 UP000183832 UP000000673 UP000069272 UP000027135 UP000242457 UP000005203 UP000053105 UP000235965 UP000007266 UP000053097 UP000279307 UP000008237 UP000076502 UP000078542 UP000075809 UP000000311 UP000078540 UP000192223 UP000030765 UP000009046 UP000053825 UP000215335 UP000092462 UP000002358 UP000094527 UP000030742
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
H9B419
Q2HZG2
H9JC27
A0A2H1WVM2
I4DJT6
A0A194R5J9
+ More
A0A0L7KV09 A0A212FG92 S4NXS5 F1AAP3 E3SWU4 E3SWV1 Q7Q8V5 A0A182U9C3 A0A182WUS9 A0A182LQ05 A0A182HFT7 A0A182VIE3 A0A182SY88 A0A1S4H4B4 A0A182K9W5 A0A1Q3FZM9 A0A1Q3FZL8 A0A1Q3FZS1 A0A1Q3FZQ3 A0A1Q3FZI2 A0A182PJK2 A0A1Q3FZI1 A0A182M3Y0 A0A182YF62 A0A1L8DNH0 A0A182VT89 A0A1L8DNC9 A0A182JE23 A0A182QKC9 N6UB80 A0A182RZ12 A0A182MYB3 A0A023EUE1 A0A182H3N7 A0A023EUI4 A0A1J1ISZ4 A0A023ETP7 A0A023EW07 A0A2M4BJ17 A0A2M4BJ01 W5JDJ6 A0A2M4ATQ7 A0A2M4BMB2 A0A2M4BLF6 A0A182FUI0 A0A290GAZ2 A0A067RKF2 A0A1Y1LM04 A0A2A3EAP0 V9IFQ1 A0A088AMI6 A0A0N0BG79 A0A2J7PLV4 D6WYJ5 D1LZL0 A0A026X0A9 E2BHH2 A0A154P303 A0A195CRN3 A0A310S9B6 E9J0G9 A0A151WRD6 E2ABX0 A0A151I4U0 A0A1W4XS25 A0A084WAU3 A0A0C9QZK4 E0VCA3 A0A0L7RF83 A0A232F2M5 A0A1B0DC19 K7IYF4 K7J772 A0A232EUG6 K7IYA1 A0A232F6B1 A0A1D2MCF9 U4TX99 N6TS19 A0A1W4XEE6 A0A290GC51 D1LZL2 D6WH81 A0A1W4X3L4 K7J7C1 A0A232EVL6 D6WH83 A0A290GSI7 D6WH82
A0A0L7KV09 A0A212FG92 S4NXS5 F1AAP3 E3SWU4 E3SWV1 Q7Q8V5 A0A182U9C3 A0A182WUS9 A0A182LQ05 A0A182HFT7 A0A182VIE3 A0A182SY88 A0A1S4H4B4 A0A182K9W5 A0A1Q3FZM9 A0A1Q3FZL8 A0A1Q3FZS1 A0A1Q3FZQ3 A0A1Q3FZI2 A0A182PJK2 A0A1Q3FZI1 A0A182M3Y0 A0A182YF62 A0A1L8DNH0 A0A182VT89 A0A1L8DNC9 A0A182JE23 A0A182QKC9 N6UB80 A0A182RZ12 A0A182MYB3 A0A023EUE1 A0A182H3N7 A0A023EUI4 A0A1J1ISZ4 A0A023ETP7 A0A023EW07 A0A2M4BJ17 A0A2M4BJ01 W5JDJ6 A0A2M4ATQ7 A0A2M4BMB2 A0A2M4BLF6 A0A182FUI0 A0A290GAZ2 A0A067RKF2 A0A1Y1LM04 A0A2A3EAP0 V9IFQ1 A0A088AMI6 A0A0N0BG79 A0A2J7PLV4 D6WYJ5 D1LZL0 A0A026X0A9 E2BHH2 A0A154P303 A0A195CRN3 A0A310S9B6 E9J0G9 A0A151WRD6 E2ABX0 A0A151I4U0 A0A1W4XS25 A0A084WAU3 A0A0C9QZK4 E0VCA3 A0A0L7RF83 A0A232F2M5 A0A1B0DC19 K7IYF4 K7J772 A0A232EUG6 K7IYA1 A0A232F6B1 A0A1D2MCF9 U4TX99 N6TS19 A0A1W4XEE6 A0A290GC51 D1LZL2 D6WH81 A0A1W4X3L4 K7J7C1 A0A232EVL6 D6WH83 A0A290GSI7 D6WH82
PDB
5YYL
E-value=1.12892e-25,
Score=290
Ontologies
PANTHER
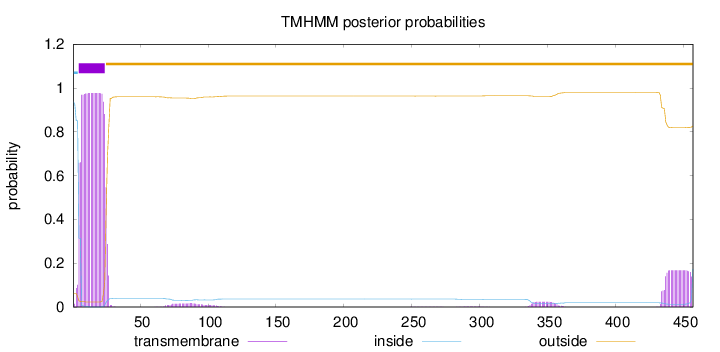
Topology
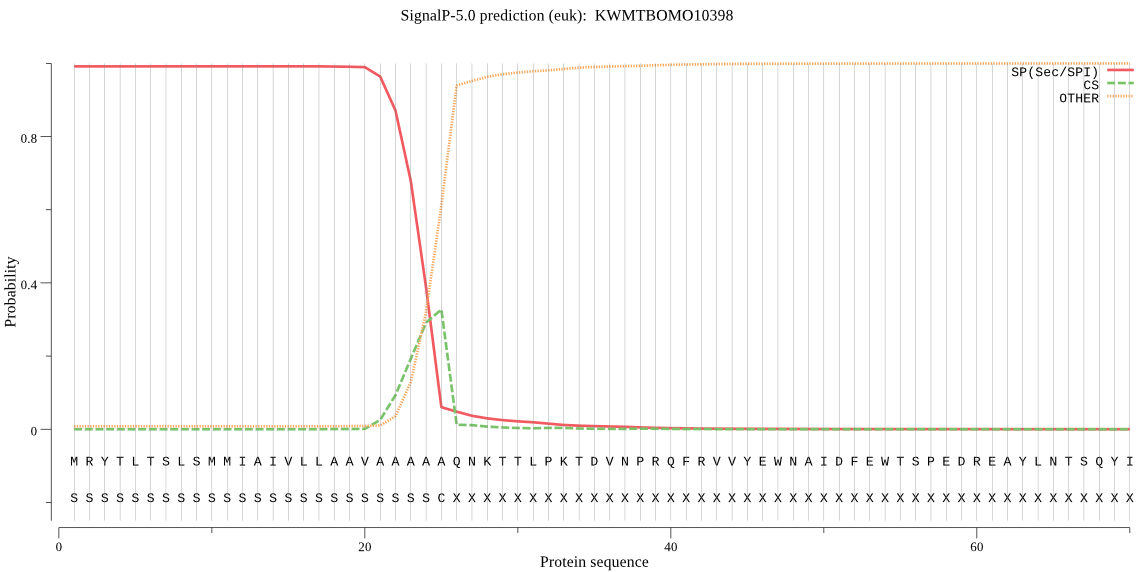
SignalP
Position: 1 - 25,
Likelihood: 0.991756
Length:
457
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
24.41201
Exp number, first 60 AAs:
19.93832
Total prob of N-in:
0.93787
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 24
outside
25 - 457
Population Genetic Test Statistics
Pi
185.402686
Theta
181.745908
Tajima's D
0.394009
CLR
0
CSRT
0.48297585120744
Interpretation
Uncertain
