Gene
KWMTBOMO10397
Pre Gene Modal
BGIBMGA007034
Annotation
PREDICTED:_G_protein-activated_inward_rectifier_potassium_channel_3-like_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.877
Sequence
CDS
ATGTCTGACAAGACCTCATCGTCGGTCGGTCCCCGAAGTTTCGAGACGATTTCGGAGGAGGACGTGACGTCACCCTGGGAGAAACTGTTGGATGATGTTAATACTGCCGTTCCTACTATAATTAAAACTAATGCGTCGAGTGCAACGATTTCAAGAATCGGGGAGGAGGTGTCTACGCACCCCCGAGTCGTGGACTTAGAGGCGAACAAGTTGACGCGCTCCAAGAGTCTCAACCAGCGGAGGAATAGCAGCGGCCTCCTCAGTTCTATCCCCCGGCAGCTGCGGCTCTCTCTCCGTGGCGTTCGCAGTGAACCCTCCAGCGTCAAGGACGTCCCGACAACTAGAAACGATAGTGCCCTCAATCTTCTTCTTAAATACCGTAATGCTCGTTACGCAGCGAGACGTGTGCGGAAGCGCGTCATCTTCAAGCACGGAGACTGCAACGTGGTCCAATGGAACGTGGCTAAGCGCAGGCGCCGATACCTTCAGGACATCTTCACGACCCTCGTGGACGCCCAGTGGCGCTGGACGCTGCTAGTGTTTGCGCTTTCCTTCATCCTCTCGTGGCTCTTGTTCGCACTTATCTGGTGGCTCATTATTTTCACTCACGGAGACCTCACCCCAATTCCACTTACGGAACAGAGAGAACCACCCGTTCCCTGCCTCAACAACGTCAACTCCTTTACTGGCTGCTTCCTCTTTTCTGTTGAAACTCAGCACACCATCGGCTATGGATCAAGGACAACGAATGAAGAATGCCCCGAAGCGATATTCGTCATGTGCATACAAAGTATTGTCGGAGTTTTCATTCAAGCTTTCATGGTTGGGATCGTGTTCGCGAAACTCTCTAGGCCAAAAAAACGCGCTCAAACTCTTCTGTACTCCAGGAACGCCGTAATTTGTTTAAGAGACGGGCAACTGTGTCTGATGTTCCGAGTCGGCGACATGAGAAAATCTCACATAGTCGAAGCTCATATCAGAGCGCAAATCATACGCCGCAAGATAACAAGGGAAGGGGAAGTTTTGCCCTTTTACCAGCAAGAGTTGAAGGTGGGAGCTGATGGCGAAGAAGACAGACTTATGTTTATTTGGCCTATGACCATAGTACACAAGATCAATGAGAAATCTCCGCTATACAATTTGTCAGCGTCGGATATGCTCAGAGAGAGATTCGAGATCGTAGTTATGCTTGAGGGTGTCATAGAGTCAACGGGAATGACAACTCAAGCCCGCAGCAGCTTCTTACCATCAGAAATTCTCTGGGGACATCGTTTCGAGACCATGGTCAAGTTCAGAAAGGATACCGGTGAATATGAGGTTGATTACACGCGGTTTAACAACACATACGAGGTGGACACGCCTTTGTGTTCAGCAAAACAACTAGACGAACTGCGCGCCTCTGTTTCAACTTCACAGAAACTCGATCGCATGCTACCGAAAACATTCTCAAACGACACCCTAGAGCTGAGCTCCATCGATTCGATGTCTTTGGATGAGCACATCGAGATTAAGATTCCGGAATCGAGGGCCCGCGAGAACAGACTGATGGCGCAAACAAACTTTGTACAACACGTCAACGAGAAAAACAAAAACCCAAGTCATCATAACCACTTGGCCGTTGAAAACGGACAAGATTTACTGCACAAGAACACTATAGAGAGTAGAGTCGAAGGAAAGGACAATCATCATATGCACAGAAGTCATAGTCATGCCAGCATGAAGAGGGTTCATGGGCTAATACCGAACGGGATATGCCATCCGATTAGTGGTCATGAACATGTACCCGACGATCATACTCTAAACAAATCCCCGAGTGAAAACTTCATAGGGAAAACTCCAGTGATTCCCATCCTTGTAACGTCACCGGAGGCGGAGCCCGCCTGA
Protein
MSDKTSSSVGPRSFETISEEDVTSPWEKLLDDVNTAVPTIIKTNASSATISRIGEEVSTHPRVVDLEANKLTRSKSLNQRRNSSGLLSSIPRQLRLSLRGVRSEPSSVKDVPTTRNDSALNLLLKYRNARYAARRVRKRVIFKHGDCNVVQWNVAKRRRRYLQDIFTTLVDAQWRWTLLVFALSFILSWLLFALIWWLIIFTHGDLTPIPLTEQREPPVPCLNNVNSFTGCFLFSVETQHTIGYGSRTTNEECPEAIFVMCIQSIVGVFIQAFMVGIVFAKLSRPKKRAQTLLYSRNAVICLRDGQLCLMFRVGDMRKSHIVEAHIRAQIIRRKITREGEVLPFYQQELKVGADGEEDRLMFIWPMTIVHKINEKSPLYNLSASDMLRERFEIVVMLEGVIESTGMTTQARSSFLPSEILWGHRFETMVKFRKDTGEYEVDYTRFNNTYEVDTPLCSAKQLDELRASVSTSQKLDRMLPKTFSNDTLELSSIDSMSLDEHIEIKIPESRARENRLMAQTNFVQHVNEKNKNPSHHNHLAVENGQDLLHKNTIESRVEGKDNHHMHRSHSHASMKRVHGLIPNGICHPISGHEHVPDDHTLNKSPSENFIGKTPVIPILVTSPEAEPA
Summary
Similarity
Belongs to the inward rectifier-type potassium channel (TC 1.A.2.1) family.
Uniprot
EMBL
GDQN01000186
JAT90868.1
GDQN01009071
JAT81983.1
BABH01014775
RSAL01000218
+ More
RVE44090.1 ODYU01001885 SOQ38720.1 KQ460685 KPJ12787.1 KQ459603 KPI93009.1 AGBW02008324 OWR53662.1 JTDY01000713 KOB76126.1 GFDF01009095 JAV04989.1 GDAI01000918 JAI16685.1 GBYB01001007 GBYB01001857 JAG70774.1 JAG71624.1 GAKP01013540 JAC45412.1 CH933806 KRG02090.1 GEZM01078207 JAV62860.1 OUUW01000005 SPP80118.1 GEZM01078196 GEZM01078195 JAV62879.1 GEZM01078206 JAV62861.1 GEZM01078203 JAV62865.1 KJ596497 AIF73577.1 GEZM01078205 JAV62862.1 CH940650 KRF83197.1 GFDL01001404 JAV33641.1
RVE44090.1 ODYU01001885 SOQ38720.1 KQ460685 KPJ12787.1 KQ459603 KPI93009.1 AGBW02008324 OWR53662.1 JTDY01000713 KOB76126.1 GFDF01009095 JAV04989.1 GDAI01000918 JAI16685.1 GBYB01001007 GBYB01001857 JAG70774.1 JAG71624.1 GAKP01013540 JAC45412.1 CH933806 KRG02090.1 GEZM01078207 JAV62860.1 OUUW01000005 SPP80118.1 GEZM01078196 GEZM01078195 JAV62879.1 GEZM01078206 JAV62861.1 GEZM01078203 JAV62865.1 KJ596497 AIF73577.1 GEZM01078205 JAV62862.1 CH940650 KRF83197.1 GFDL01001404 JAV33641.1
Proteomes
PRIDE
Interpro
SUPFAM
SSF81296
SSF81296
Gene 3D
ProteinModelPortal
PDB
3SPI
E-value=1.36229e-91,
Score=860
Ontologies
GO
PANTHER
Topology
Subcellular location
Membrane
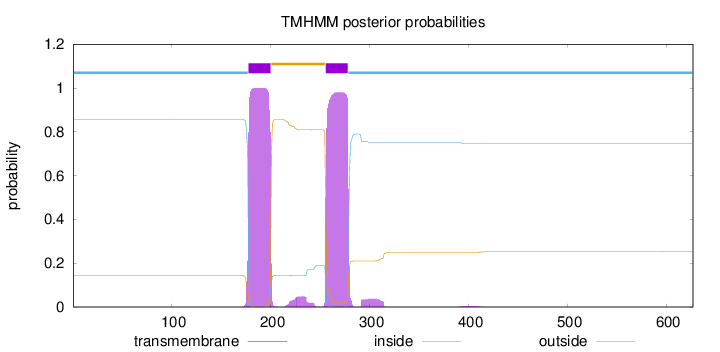
Length:
627
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
47.29982
Exp number, first 60 AAs:
0.0016
Total prob of N-in:
0.85629
inside
1 - 177
TMhelix
178 - 200
outside
201 - 255
TMhelix
256 - 278
inside
279 - 627
Population Genetic Test Statistics
Pi
281.206716
Theta
194.749281
Tajima's D
0.629665
CLR
0.467515
CSRT
0.556922153892305
Interpretation
Uncertain
