Gene
KWMTBOMO10396
Pre Gene Modal
BGIBMGA007070
Annotation
PREDICTED:_G_protein-activated_inward_rectifier_potassium_channel_3-like_isoform_X3_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 3.422
Sequence
CDS
ATGAGAGTGTCTGTCAGTCGTGACCGAAATATTATCAGGCGCGATGCTTCAGGAGGCTTGAGATCATCGAGGGATGGTGGCGCAGACAGTACGCGATTGAGACGCGCGCACGACGCCTGCGCACCAGACTCCGCCATGCACGCCGATCCCGAGTCGCCCTGTTGTCGTGAGGAGACACAGCTGCTCCCCGACGAGTGGCTCAAAGTCAAGACGGTACCCACCAACGGGAACCTATCGCCCGGAGTCTACTATCCAACAGGCAGCAAGAGGAGCCGATCGCACCGGCATGGCGTCACTAGAAGGACTAGACGGAGAGCGATTCTCAAAAATGGAGAGTGCAATATTCTCAAGTCCCGAATCTCTCAGCGGCGTTTGAGATTCCTGCAAGACATGTTTACGACTTTAGTTGACGCACAATGGCGGTGGACCCTCCTTGTTTTCACACTATCGTTTATACTGTCGTGGTTGGGCTTTGCGTTGATCTGGTGGCTGATAGCATTCACACACGGCGACCTGGAACCGGACCACCTACCGCCATTACAGGACTCTGCGAATTGGAAGCCGTGCGTTTTTAACATAAACGATTTCACGTCGTGTTTCTTGTTCAGTATCGAAACGCAGCACACCATTGGCTACGGTGCCCGTACTACAACGGAAGAATGTCCGGAGGCCATTTTCATAATGTGCCTCCAAAGTATCGTCGGTGTGATGATTCAGGCGTTCATGGTCGGTATCGTGTTCGCAAAGATGACAAGACCTAAGCATAGAACGCAGACGTTACTGTTCTCGAAGTATGCAGTCGTGTGTCAAAGAGATGGCGAGTTAAGTTTAATGTTCCGAGTGGGCGACTTGAGGAAATCGCACATAATAGGGGCGAGCGTGAGAGCGCAGTTGATTCGATCGCGGACGACTAAGGAAGGAGAGGTATTGTCGCATTATCAAACCGAATTAGAACTCAACGCAGATGGATGTGACAGCAATCTTTTCTTCATCTGGCCTATTACGATGGTACATAGAATAAATCGCGACAGCCCCTTCTACGGAGTGTCAGCGGCGGACGTACTACAAGAAAGATTCGAAATAGTAGTTATATTGGAAGGAACAATTGAATCGACGGGTCAAACCACACAGGCGCGCTCCAGTTACACAACCAGTGAAATTATGTGGGGGCATAGGTTCGTTCCGCTTGTGACGTACAACCGTGAACGACAAGGATATCAAGTGGATTATTCTAAATTTGACGAGACGACCCAGGTGGACACTCCGCTCTGTTCAGCAAAAGAGCTAGACGAATTTTACGGCAGCCAGGCTGATCGTAGATCTATCGAAAGCGCGGAACATCCACAGCAGCAGTTAATTCTGAAGATGCCGGATAGAGCTCCTACCACCGAACCTAAGGCCGAAAATAAAGACGGCGACTTCACTGTTACGTTGTAA
Protein
MRVSVSRDRNIIRRDASGGLRSSRDGGADSTRLRRAHDACAPDSAMHADPESPCCREETQLLPDEWLKVKTVPTNGNLSPGVYYPTGSKRSRSHRHGVTRRTRRRAILKNGECNILKSRISQRRLRFLQDMFTTLVDAQWRWTLLVFTLSFILSWLGFALIWWLIAFTHGDLEPDHLPPLQDSANWKPCVFNINDFTSCFLFSIETQHTIGYGARTTTEECPEAIFIMCLQSIVGVMIQAFMVGIVFAKMTRPKHRTQTLLFSKYAVVCQRDGELSLMFRVGDLRKSHIIGASVRAQLIRSRTTKEGEVLSHYQTELELNADGCDSNLFFIWPITMVHRINRDSPFYGVSAADVLQERFEIVVILEGTIESTGQTTQARSSYTTSEIMWGHRFVPLVTYNRERQGYQVDYSKFDETTQVDTPLCSAKELDEFYGSQADRRSIESAEHPQQQLILKMPDRAPTTEPKAENKDGDFTVTL
Summary
Similarity
Belongs to the inward rectifier-type potassium channel (TC 1.A.2.1) family.
Uniprot
A0A194PIY2
H9JC25
A0A212FIV9
A0A2H1VCZ2
A0A0L7LKP4
A0A194RA99
+ More
A0A1E1W6K4 A0A3S2NCQ4 A0A1Q3FNY2 B0XCY9 A0A1Q3FNU5 A0A1S4FL01 Q16XB8 A0A1Q3FUZ4 A0A1Q3FUB8 Q16XB6 Q16XB7 A0A023ES54 A0A1Q3G4Y4 U5EGP0 A0A1Q3G4W8 A0A0L7REX2 A0A1J1IE18 A0A1J1IHJ0 V5G7T8 N6T150 E2BHE4 A0A0N0U587 J3JV45 A0A1Y1NIH2 A0A1Y1NJY8 A0A3L8DEV9 E9IHQ4 A0A158NMU5 A0A182QA82 E2A7T2 A0A195FXP7 V9I7I7 A0A088AR70 A0A026WAU9 A0A151X0B2 A0A151IS07 A0A195C1D6 A0A0C9PH29 A0A182VGL1 A0A1Y9J109 A0A1Y9J0B9 A0A2J7Q3L9 A0A310S9C3 A7UVQ1 A0A1Y9J115 A0A1Y9J0K5 A0A067QNF1 A0A1C7CZS3 A0A182UKL8 A0A2C9GQR0 Q7PKK8 A0A2J7Q3M0 A0A0C9RAJ9 A0A2M4AH79 A0A2M4CNC0 W5JA40 A0A0T6AWB8 A0A182M6E0 A0A1Y9IV43 A0A1L8DF16 A0A2M4BN11 A0A2M4CNA8 A0A1Y9IVK2 A0A084W0M9 A0A232EUK4 A0A182J0B7 A0A2M3ZH10 A0A2M4BN81 T1EA29 A0A2M4BNE8 A0A182JDE4 A0A2M4BR23 A0A2M3Z3W1 F5HMU3 A0A0K8WIH1 A0A1Y9J120 A0A084W0N0 A0A0A1X5V3 A0A0K8V2R8 A0A182TWH8 A0A182TA50 A0A182MQE6 A0A0K8UWX1 A0A1Y9IVW0 A0A182V716 A0A182L8Q7 A0A2C9GQH5 Q7PX91 A0A0A1XQR2 A0A1I8NYM8 W8BA85
A0A1E1W6K4 A0A3S2NCQ4 A0A1Q3FNY2 B0XCY9 A0A1Q3FNU5 A0A1S4FL01 Q16XB8 A0A1Q3FUZ4 A0A1Q3FUB8 Q16XB6 Q16XB7 A0A023ES54 A0A1Q3G4Y4 U5EGP0 A0A1Q3G4W8 A0A0L7REX2 A0A1J1IE18 A0A1J1IHJ0 V5G7T8 N6T150 E2BHE4 A0A0N0U587 J3JV45 A0A1Y1NIH2 A0A1Y1NJY8 A0A3L8DEV9 E9IHQ4 A0A158NMU5 A0A182QA82 E2A7T2 A0A195FXP7 V9I7I7 A0A088AR70 A0A026WAU9 A0A151X0B2 A0A151IS07 A0A195C1D6 A0A0C9PH29 A0A182VGL1 A0A1Y9J109 A0A1Y9J0B9 A0A2J7Q3L9 A0A310S9C3 A7UVQ1 A0A1Y9J115 A0A1Y9J0K5 A0A067QNF1 A0A1C7CZS3 A0A182UKL8 A0A2C9GQR0 Q7PKK8 A0A2J7Q3M0 A0A0C9RAJ9 A0A2M4AH79 A0A2M4CNC0 W5JA40 A0A0T6AWB8 A0A182M6E0 A0A1Y9IV43 A0A1L8DF16 A0A2M4BN11 A0A2M4CNA8 A0A1Y9IVK2 A0A084W0M9 A0A232EUK4 A0A182J0B7 A0A2M3ZH10 A0A2M4BN81 T1EA29 A0A2M4BNE8 A0A182JDE4 A0A2M4BR23 A0A2M3Z3W1 F5HMU3 A0A0K8WIH1 A0A1Y9J120 A0A084W0N0 A0A0A1X5V3 A0A0K8V2R8 A0A182TWH8 A0A182TA50 A0A182MQE6 A0A0K8UWX1 A0A1Y9IVW0 A0A182V716 A0A182L8Q7 A0A2C9GQH5 Q7PX91 A0A0A1XQR2 A0A1I8NYM8 W8BA85
Pubmed
EMBL
KQ459603
KPI93008.1
BABH01014775
AGBW02008324
OWR53664.1
ODYU01001885
+ More
SOQ38719.1 JTDY01000713 KOB76123.1 KQ460685 KPJ12786.1 GDQN01008428 JAT82626.1 RSAL01000218 RVE44089.1 GFDL01005823 JAV29222.1 DS232728 EDS45184.1 GFDL01005841 JAV29204.1 KF686746 CH477543 AIN76714.1 EAT39258.1 GFDL01003615 JAV31430.1 GFDL01003851 JAV31194.1 KF686745 AIN76713.1 EAT39257.1 KF686744 AIN76712.1 EAT39256.1 GAPW01001525 JAC12073.1 GFDL01000192 JAV34853.1 GANO01003364 JAB56507.1 GFDL01000195 JAV34850.1 KQ414608 KOC69364.1 CVRI01000047 CRK98501.1 CRK98502.1 GALX01002281 JAB66185.1 APGK01057039 KB741277 ENN71258.1 GL448287 EFN84913.1 KQ435791 KOX74402.1 BT127111 KB630626 KB631617 KB631850 AEE62073.1 ERL83730.1 ERL84721.1 ERL86721.1 GEZM01004174 JAV96445.1 GEZM01004173 JAV96446.1 QOIP01000009 RLU18987.1 GL763289 EFZ19901.1 ADTU01002433 ADTU01002434 ADTU01002435 ADTU01002436 ADTU01002437 AXCN02001514 GL437384 EFN70512.1 KQ981200 KYN45082.1 JR035661 JR035662 JR035663 JR035664 JR035665 JR035666 JR035667 JR035668 JR035669 JR035670 JR035671 JR035672 AEY56573.1 AEY56574.1 AEY56575.1 AEY56576.1 AEY56577.1 AEY56578.1 AEY56579.1 AEY56580.1 AEY56581.1 AEY56582.1 AEY56583.1 AEY56584.1 KK107320 EZA52801.1 KQ982617 KYQ53796.1 KQ981113 KYN09394.1 KQ978350 KYM94689.1 GBYB01000163 JAG69930.1 NEVH01019065 PNF23184.1 KQ767621 OAD53388.1 AAAB01008987 EDO63275.2 KK853134 KDR10854.1 APCN01000829 EAA43222.5 PNF23185.1 GBYB01003841 JAG73608.1 GGFK01006647 MBW39968.1 GGFL01002636 MBW66814.1 ADMH02002066 ETN59720.1 LJIG01022677 KRT79300.1 AXCM01007933 GFDF01009053 JAV05031.1 GGFJ01005329 MBW54470.1 GGFL01002581 MBW66759.1 ATLV01019123 KE525262 KFB43773.1 NNAY01002130 OXU21996.1 GGFM01007085 MBW27836.1 GGFJ01005331 MBW54472.1 GAMD01000792 JAB00799.1 GGFJ01005330 MBW54471.1 GGFJ01006368 MBW55509.1 GGFM01002458 MBW23209.1 EGK97615.1 GDHF01001363 JAI50951.1 KFB43774.1 GBXI01007785 JAD06507.1 GDHF01019449 JAI32865.1 GDHF01021228 JAI31086.1 EAA01723.5 GBXI01001394 JAD12898.1 GAMC01019801 JAB86754.1
SOQ38719.1 JTDY01000713 KOB76123.1 KQ460685 KPJ12786.1 GDQN01008428 JAT82626.1 RSAL01000218 RVE44089.1 GFDL01005823 JAV29222.1 DS232728 EDS45184.1 GFDL01005841 JAV29204.1 KF686746 CH477543 AIN76714.1 EAT39258.1 GFDL01003615 JAV31430.1 GFDL01003851 JAV31194.1 KF686745 AIN76713.1 EAT39257.1 KF686744 AIN76712.1 EAT39256.1 GAPW01001525 JAC12073.1 GFDL01000192 JAV34853.1 GANO01003364 JAB56507.1 GFDL01000195 JAV34850.1 KQ414608 KOC69364.1 CVRI01000047 CRK98501.1 CRK98502.1 GALX01002281 JAB66185.1 APGK01057039 KB741277 ENN71258.1 GL448287 EFN84913.1 KQ435791 KOX74402.1 BT127111 KB630626 KB631617 KB631850 AEE62073.1 ERL83730.1 ERL84721.1 ERL86721.1 GEZM01004174 JAV96445.1 GEZM01004173 JAV96446.1 QOIP01000009 RLU18987.1 GL763289 EFZ19901.1 ADTU01002433 ADTU01002434 ADTU01002435 ADTU01002436 ADTU01002437 AXCN02001514 GL437384 EFN70512.1 KQ981200 KYN45082.1 JR035661 JR035662 JR035663 JR035664 JR035665 JR035666 JR035667 JR035668 JR035669 JR035670 JR035671 JR035672 AEY56573.1 AEY56574.1 AEY56575.1 AEY56576.1 AEY56577.1 AEY56578.1 AEY56579.1 AEY56580.1 AEY56581.1 AEY56582.1 AEY56583.1 AEY56584.1 KK107320 EZA52801.1 KQ982617 KYQ53796.1 KQ981113 KYN09394.1 KQ978350 KYM94689.1 GBYB01000163 JAG69930.1 NEVH01019065 PNF23184.1 KQ767621 OAD53388.1 AAAB01008987 EDO63275.2 KK853134 KDR10854.1 APCN01000829 EAA43222.5 PNF23185.1 GBYB01003841 JAG73608.1 GGFK01006647 MBW39968.1 GGFL01002636 MBW66814.1 ADMH02002066 ETN59720.1 LJIG01022677 KRT79300.1 AXCM01007933 GFDF01009053 JAV05031.1 GGFJ01005329 MBW54470.1 GGFL01002581 MBW66759.1 ATLV01019123 KE525262 KFB43773.1 NNAY01002130 OXU21996.1 GGFM01007085 MBW27836.1 GGFJ01005331 MBW54472.1 GAMD01000792 JAB00799.1 GGFJ01005330 MBW54471.1 GGFJ01006368 MBW55509.1 GGFM01002458 MBW23209.1 EGK97615.1 GDHF01001363 JAI50951.1 KFB43774.1 GBXI01007785 JAD06507.1 GDHF01019449 JAI32865.1 GDHF01021228 JAI31086.1 EAA01723.5 GBXI01001394 JAD12898.1 GAMC01019801 JAB86754.1
Proteomes
UP000053268
UP000005204
UP000007151
UP000037510
UP000053240
UP000283053
+ More
UP000002320 UP000008820 UP000053825 UP000183832 UP000019118 UP000008237 UP000053105 UP000030742 UP000279307 UP000005205 UP000075886 UP000000311 UP000078541 UP000005203 UP000053097 UP000075809 UP000078492 UP000078542 UP000075903 UP000076407 UP000235965 UP000007062 UP000027135 UP000075882 UP000075902 UP000075840 UP000000673 UP000075883 UP000075920 UP000030765 UP000215335 UP000075880 UP000075901 UP000095300
UP000002320 UP000008820 UP000053825 UP000183832 UP000019118 UP000008237 UP000053105 UP000030742 UP000279307 UP000005205 UP000075886 UP000000311 UP000078541 UP000005203 UP000053097 UP000075809 UP000078492 UP000078542 UP000075903 UP000076407 UP000235965 UP000007062 UP000027135 UP000075882 UP000075902 UP000075840 UP000000673 UP000075883 UP000075920 UP000030765 UP000215335 UP000075880 UP000075901 UP000095300
PRIDE
Interpro
SUPFAM
SSF81296
SSF81296
Gene 3D
ProteinModelPortal
A0A194PIY2
H9JC25
A0A212FIV9
A0A2H1VCZ2
A0A0L7LKP4
A0A194RA99
+ More
A0A1E1W6K4 A0A3S2NCQ4 A0A1Q3FNY2 B0XCY9 A0A1Q3FNU5 A0A1S4FL01 Q16XB8 A0A1Q3FUZ4 A0A1Q3FUB8 Q16XB6 Q16XB7 A0A023ES54 A0A1Q3G4Y4 U5EGP0 A0A1Q3G4W8 A0A0L7REX2 A0A1J1IE18 A0A1J1IHJ0 V5G7T8 N6T150 E2BHE4 A0A0N0U587 J3JV45 A0A1Y1NIH2 A0A1Y1NJY8 A0A3L8DEV9 E9IHQ4 A0A158NMU5 A0A182QA82 E2A7T2 A0A195FXP7 V9I7I7 A0A088AR70 A0A026WAU9 A0A151X0B2 A0A151IS07 A0A195C1D6 A0A0C9PH29 A0A182VGL1 A0A1Y9J109 A0A1Y9J0B9 A0A2J7Q3L9 A0A310S9C3 A7UVQ1 A0A1Y9J115 A0A1Y9J0K5 A0A067QNF1 A0A1C7CZS3 A0A182UKL8 A0A2C9GQR0 Q7PKK8 A0A2J7Q3M0 A0A0C9RAJ9 A0A2M4AH79 A0A2M4CNC0 W5JA40 A0A0T6AWB8 A0A182M6E0 A0A1Y9IV43 A0A1L8DF16 A0A2M4BN11 A0A2M4CNA8 A0A1Y9IVK2 A0A084W0M9 A0A232EUK4 A0A182J0B7 A0A2M3ZH10 A0A2M4BN81 T1EA29 A0A2M4BNE8 A0A182JDE4 A0A2M4BR23 A0A2M3Z3W1 F5HMU3 A0A0K8WIH1 A0A1Y9J120 A0A084W0N0 A0A0A1X5V3 A0A0K8V2R8 A0A182TWH8 A0A182TA50 A0A182MQE6 A0A0K8UWX1 A0A1Y9IVW0 A0A182V716 A0A182L8Q7 A0A2C9GQH5 Q7PX91 A0A0A1XQR2 A0A1I8NYM8 W8BA85
A0A1E1W6K4 A0A3S2NCQ4 A0A1Q3FNY2 B0XCY9 A0A1Q3FNU5 A0A1S4FL01 Q16XB8 A0A1Q3FUZ4 A0A1Q3FUB8 Q16XB6 Q16XB7 A0A023ES54 A0A1Q3G4Y4 U5EGP0 A0A1Q3G4W8 A0A0L7REX2 A0A1J1IE18 A0A1J1IHJ0 V5G7T8 N6T150 E2BHE4 A0A0N0U587 J3JV45 A0A1Y1NIH2 A0A1Y1NJY8 A0A3L8DEV9 E9IHQ4 A0A158NMU5 A0A182QA82 E2A7T2 A0A195FXP7 V9I7I7 A0A088AR70 A0A026WAU9 A0A151X0B2 A0A151IS07 A0A195C1D6 A0A0C9PH29 A0A182VGL1 A0A1Y9J109 A0A1Y9J0B9 A0A2J7Q3L9 A0A310S9C3 A7UVQ1 A0A1Y9J115 A0A1Y9J0K5 A0A067QNF1 A0A1C7CZS3 A0A182UKL8 A0A2C9GQR0 Q7PKK8 A0A2J7Q3M0 A0A0C9RAJ9 A0A2M4AH79 A0A2M4CNC0 W5JA40 A0A0T6AWB8 A0A182M6E0 A0A1Y9IV43 A0A1L8DF16 A0A2M4BN11 A0A2M4CNA8 A0A1Y9IVK2 A0A084W0M9 A0A232EUK4 A0A182J0B7 A0A2M3ZH10 A0A2M4BN81 T1EA29 A0A2M4BNE8 A0A182JDE4 A0A2M4BR23 A0A2M3Z3W1 F5HMU3 A0A0K8WIH1 A0A1Y9J120 A0A084W0N0 A0A0A1X5V3 A0A0K8V2R8 A0A182TWH8 A0A182TA50 A0A182MQE6 A0A0K8UWX1 A0A1Y9IVW0 A0A182V716 A0A182L8Q7 A0A2C9GQH5 Q7PX91 A0A0A1XQR2 A0A1I8NYM8 W8BA85
PDB
3SPI
E-value=2.15543e-99,
Score=926
Ontologies
GO
PANTHER
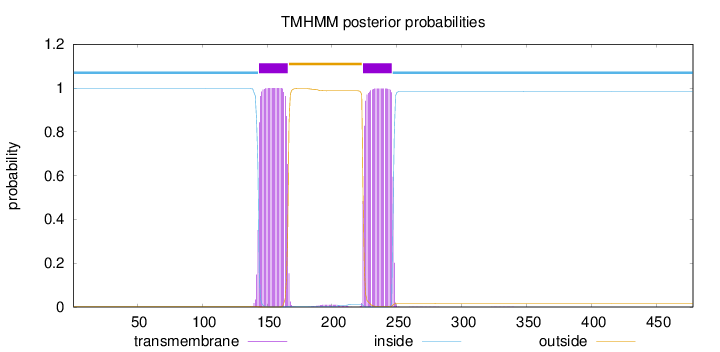
Topology
Subcellular location
Membrane
Length:
478
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
46.1505000000003
Exp number, first 60 AAs:
0
Total prob of N-in:
0.99733
inside
1 - 143
TMhelix
144 - 166
outside
167 - 223
TMhelix
224 - 246
inside
247 - 478
Population Genetic Test Statistics
Pi
262.448885
Theta
219.045867
Tajima's D
0.807667
CLR
0.002938
CSRT
0.603169841507925
Interpretation
Uncertain
