Gene
KWMTBOMO10389
Pre Gene Modal
BGIBMGA007038
Annotation
PREDICTED:_sodium-independent_sulfate_anion_transporter-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.875
Sequence
CDS
ATGTATGGCCTGTACGCGTCTTTCGTGGGTGGATTCGTTTACGCGATCATGGGCACATGCCCTCAGATCAACATTGGACCGACGGCGTTACTGTCACTTCTCACCTTCACTTACACAAGCGGCACTAACGCCGACTTCGCCGTGCTACTATGTTTCATTGCCGGAATAGTTCAGCTATTCGCTGGATTCGCACAGCTGGGGTTCTTAGTGGAGTTCGTCTCCCTACCGGTAGTGTCAGGTTTCACGTCGGCAGCTGCCATCACTATAGCTTCATCCCAAATTAAAGGCTTGTTAGGATTGAAGTTCAGCGCAGAGACCTTCATATCGACTTGGAAGGAACTCTTTGTTCATATTGGAGAAACGAGGATGCAGGACACATTACTCAGCTTATTCTGCTGCATTGTGCTGATGGGAATGAAGGCATTAAAAGATATCGAAATCAAAAGTGTAATAAACGACAAGAACCGAAAAGGTATGCTGATACTGCAAAGGTTTTTGTGGTTCGTTGGAGTCTCACGGAACGCCGTGGTCGTGGTGCTAGCTGCAGTACTGGCGTACACGGTACATGAAGATAAAAGGGACCCGCTCATATTGACGGGTAGCATTACTCCAGGCCTTCCAATTCCTCAAGCACCTGCCTTCCACACAACGGTGGGTAATACTACATATACTGCGGTGGAAATGTTCACGCATTTGGGCTCTGGTCTCTTGGTGGTTCCATTAGTTGGAATCATCTCGAATGTGGCCATCGCCAAAGCTTTCTCGAACGGCAGAACGTTGGACGCCACTCAGGAGATAGTGGCTCTGGGCGCTTGCAACTTGTTCGGGGCATTCTTTCGTTCGTTTCCCGTGAACGGTTCGTTCACACGGAGCGCTGTCAGCGAAGCTTCCGGGGTCAAGACGCCGGCCGCTGGTTTCTATACTGGTGTAATCGTATTACTGACGTTGGCCCTCCTGACTCCGTACTTCTTCTTCATTCCGCGCGCCGCGCTCTCCGCCGTGATAGTTTGCGCCGTCCTGCACATGGTCGACACGGACATACTGAAGAAACTCTGGAACACCAGCCGATTAGACCTGATACCGCTGTTGGGTTCCTTCGTATGCTGTCTAGCATTGGGTATCGAAGTGGGATTGATATGTGGAGTTATAGTAGACATGCTACTGCTGCTCTACTACCATTCCAGACCCCCACTCGATATAGTTTATGTTAATGACGGAGTTTTGCCAGCGCATTTTGCGATCCATCCGTGCGGCGGTTTGTATTTCGCTGGAGCAGAACAAGTCAGATCAAAACTGATATCTCTGAGAAAACCTAAAAACAACAGCATCCCGGCTACAGCACTAGAAACATTAACAATAATACCTGATTCGGATAATAATGGAACTAGTGATGGGAACAATATCGCTAATGGAAATCATATTGCCAACGGGAATTTATCCAATGGGAATAATAATGCCAACGGGAATAATGTTACCAATGGGAATATCAACGGTAACTTTATAAGGTCCACAGAACTTCTAGTCGTGTATTGTGACGCAGTGTCTAGAATGGACTATACTTTTTTGCAGAGTTTGAAAATGTTAGTGGCGGAATGGTCACGTGGCGGTCGCGTTTTCTGGTGTGACGCGAATCCCTTAGTGAAAGAACAATTGCTGAACGTTCTCCAGGACCCTCACTTCTGCGACGCCGAGCAGCTCAGGATACTGTTGGCCGGTACTCTGTCGGAAGATTCACAGGTAAACATAAGCGATACACAACTCTAG
Protein
MYGLYASFVGGFVYAIMGTCPQINIGPTALLSLLTFTYTSGTNADFAVLLCFIAGIVQLFAGFAQLGFLVEFVSLPVVSGFTSAAAITIASSQIKGLLGLKFSAETFISTWKELFVHIGETRMQDTLLSLFCCIVLMGMKALKDIEIKSVINDKNRKGMLILQRFLWFVGVSRNAVVVVLAAVLAYTVHEDKRDPLILTGSITPGLPIPQAPAFHTTVGNTTYTAVEMFTHLGSGLLVVPLVGIISNVAIAKAFSNGRTLDATQEIVALGACNLFGAFFRSFPVNGSFTRSAVSEASGVKTPAAGFYTGVIVLLTLALLTPYFFFIPRAALSAVIVCAVLHMVDTDILKKLWNTSRLDLIPLLGSFVCCLALGIEVGLICGVIVDMLLLLYYHSRPPLDIVYVNDGVLPAHFAIHPCGGLYFAGAEQVRSKLISLRKPKNNSIPATALETLTIIPDSDNNGTSDGNNIANGNHIANGNLSNGNNNANGNNVTNGNINGNFIRSTELLVVYCDAVSRMDYTFLQSLKMLVAEWSRGGRVFWCDANPLVKEQLLNVLQDPHFCDAEQLRILLAGTLSEDSQVNISDTQL
Summary
Similarity
Belongs to the SLC26A/SulP transporter (TC 2.A.53) family.
Uniprot
H9JBZ3
A0A194PIW7
A0A0N1PJT0
A0A3S2LU88
A0A1W4XDG5
A0A2J7RSL0
+ More
A0A2A3EE05 A0A1B6MPW8 E0VCB0 A0A3L8DVY2 V9I9F7 A0A154PRJ3 A0A1W4XEP4 E2AWV5 A0A1W4XEN4 A0A1W4XEM9 A0A1B6CML3 A0A1Y1MPJ6 A0A195FJ73 A0A2J7QAB9 A0A2J7QAE0 A0A067RPK8 A0A1B6C3Z6 A0A139WD39 A0A069DWI2 A0A088AR89 A0A2A3EBU0 A0A1B6EVA9 A0A023F3H2 A0A3Q0J7I5 E2BSP6 A0A0P4VQI8 T1IES7 A0A069DV45 A0A023F3Y6 A0A224X9U9 A0A139WCX6 F4WUD0 A0A151J6H4 A0A195BVT9 H9JBZ6 A0A0L7RCE8 A0A151I5C0 A0A158NL57 E2ANG6 A0A154PR24 A0A194PIX2 A0A158NTY8 A0A0M9A911 A0A151X835 A0A195FKC5 A0A2S2NQ78 D2A570 T1HT78 K7IX01 A0A0T6BGS6 K7IYL9 A0A195C7D8 J9JIX9 A0A026X0F7 A0A0C9Q9Z3 A0A2W1BKM4 A0A151I4S0 A0A158NTY7 A0A2J7RSK1 A0A0T6B2L2 A0A2J7PKW2 A0A212F938 A0A232FF55 A0A1B6BYH8 A0A0A9XS76 A0A1W4X674 A0A2A4IZ56 A0A212FP71 A0A2H1V6V8 A0A0A9Z6Y1 A0A0V0G6Q9 A0A146KSN0 A0A1B0D6L8 A0A0P4WEK6 A0A0A9Z2J6 A0A146M619 A0A0P4WSY8 A0A195BU35 A0A0P4W8Z7 A0A154P5U0 E0VH04
A0A2A3EE05 A0A1B6MPW8 E0VCB0 A0A3L8DVY2 V9I9F7 A0A154PRJ3 A0A1W4XEP4 E2AWV5 A0A1W4XEN4 A0A1W4XEM9 A0A1B6CML3 A0A1Y1MPJ6 A0A195FJ73 A0A2J7QAB9 A0A2J7QAE0 A0A067RPK8 A0A1B6C3Z6 A0A139WD39 A0A069DWI2 A0A088AR89 A0A2A3EBU0 A0A1B6EVA9 A0A023F3H2 A0A3Q0J7I5 E2BSP6 A0A0P4VQI8 T1IES7 A0A069DV45 A0A023F3Y6 A0A224X9U9 A0A139WCX6 F4WUD0 A0A151J6H4 A0A195BVT9 H9JBZ6 A0A0L7RCE8 A0A151I5C0 A0A158NL57 E2ANG6 A0A154PR24 A0A194PIX2 A0A158NTY8 A0A0M9A911 A0A151X835 A0A195FKC5 A0A2S2NQ78 D2A570 T1HT78 K7IX01 A0A0T6BGS6 K7IYL9 A0A195C7D8 J9JIX9 A0A026X0F7 A0A0C9Q9Z3 A0A2W1BKM4 A0A151I4S0 A0A158NTY7 A0A2J7RSK1 A0A0T6B2L2 A0A2J7PKW2 A0A212F938 A0A232FF55 A0A1B6BYH8 A0A0A9XS76 A0A1W4X674 A0A2A4IZ56 A0A212FP71 A0A2H1V6V8 A0A0A9Z6Y1 A0A0V0G6Q9 A0A146KSN0 A0A1B0D6L8 A0A0P4WEK6 A0A0A9Z2J6 A0A146M619 A0A0P4WSY8 A0A195BU35 A0A0P4W8Z7 A0A154P5U0 E0VH04
Pubmed
EMBL
BABH01014760
KQ459603
KPI92993.1
KQ460328
KPJ15650.1
RSAL01000218
+ More
RVE44086.1 NEVH01000254 PNF43821.1 KZ288289 PBC29241.1 GEBQ01002062 JAT37915.1 DS235051 EEB10996.1 QOIP01000004 RLU23928.1 JR037370 AEY57743.1 KQ435010 KZC13730.1 GL443425 EFN62075.1 GEDC01022690 JAS14608.1 GEZM01025131 JAV87571.1 KQ981522 KYN40710.1 NEVH01016331 PNF25526.1 PNF25529.1 KK852498 KDR22565.1 GEDC01029081 GEDC01027874 JAS08217.1 JAS09424.1 KQ971362 KYB25751.1 GBGD01000832 JAC88057.1 PBC29243.1 GECZ01027882 JAS41887.1 GBBI01002876 JAC15836.1 GL450241 EFN81292.1 GDKW01003168 JAI53427.1 ACPB03015825 GBGD01000966 JAC87923.1 GBBI01002547 JAC16165.1 GFTR01007289 JAW09137.1 KYB25752.1 GL888355 EGI62238.1 KQ979854 KYN18808.1 KQ976403 KYM92093.1 BABH01014720 BABH01014721 KQ414615 KOC68513.1 KQ976445 KYM86091.1 ADTU01002138 ADTU01002139 ADTU01002140 GL441216 EFN65015.1 KZC13728.1 KPI92998.1 ADTU01026211 ADTU01026212 KQ435711 KOX79687.1 KQ982431 KYQ56537.1 KYN40712.1 GGMR01006712 MBY19331.1 KQ971361 EFA05122.1 ACPB03016174 AAZX01007293 LJIG01000412 KRT86528.1 KQ978143 KYM96789.1 ABLF02031674 ABLF02031675 ABLF02031682 ABLF02057042 KK107046 EZA61752.1 RLU23357.1 GBYB01011218 GBYB01011219 JAG80985.1 JAG80986.1 KZ149989 PZC75632.1 KYM86093.1 ADTU01026210 PNF43809.1 LJIG01016073 KRT81695.1 NEVH01024531 PNF16976.1 AGBW02009651 OWR50240.1 NNAY01000295 OXU29396.1 GEDC01030992 JAS06306.1 GBHO01021951 JAG21653.1 NWSH01004565 PCG64919.1 AGBW02003464 OWR55546.1 ODYU01000799 SOQ36122.1 GBHO01006039 JAG37565.1 GECL01002483 JAP03641.1 GDHC01019650 JAP98978.1 AJVK01012081 AJVK01012082 GDRN01063798 JAI64926.1 GBHO01006041 GBHO01006040 GBRD01002176 JAG37563.1 JAG37564.1 JAG63645.1 GDHC01003426 JAQ15203.1 GDRN01063797 JAI64927.1 KYM92092.1 GDRN01063799 JAI64925.1 KQ434823 KZC07237.1 DS235156 EEB12660.1
RVE44086.1 NEVH01000254 PNF43821.1 KZ288289 PBC29241.1 GEBQ01002062 JAT37915.1 DS235051 EEB10996.1 QOIP01000004 RLU23928.1 JR037370 AEY57743.1 KQ435010 KZC13730.1 GL443425 EFN62075.1 GEDC01022690 JAS14608.1 GEZM01025131 JAV87571.1 KQ981522 KYN40710.1 NEVH01016331 PNF25526.1 PNF25529.1 KK852498 KDR22565.1 GEDC01029081 GEDC01027874 JAS08217.1 JAS09424.1 KQ971362 KYB25751.1 GBGD01000832 JAC88057.1 PBC29243.1 GECZ01027882 JAS41887.1 GBBI01002876 JAC15836.1 GL450241 EFN81292.1 GDKW01003168 JAI53427.1 ACPB03015825 GBGD01000966 JAC87923.1 GBBI01002547 JAC16165.1 GFTR01007289 JAW09137.1 KYB25752.1 GL888355 EGI62238.1 KQ979854 KYN18808.1 KQ976403 KYM92093.1 BABH01014720 BABH01014721 KQ414615 KOC68513.1 KQ976445 KYM86091.1 ADTU01002138 ADTU01002139 ADTU01002140 GL441216 EFN65015.1 KZC13728.1 KPI92998.1 ADTU01026211 ADTU01026212 KQ435711 KOX79687.1 KQ982431 KYQ56537.1 KYN40712.1 GGMR01006712 MBY19331.1 KQ971361 EFA05122.1 ACPB03016174 AAZX01007293 LJIG01000412 KRT86528.1 KQ978143 KYM96789.1 ABLF02031674 ABLF02031675 ABLF02031682 ABLF02057042 KK107046 EZA61752.1 RLU23357.1 GBYB01011218 GBYB01011219 JAG80985.1 JAG80986.1 KZ149989 PZC75632.1 KYM86093.1 ADTU01026210 PNF43809.1 LJIG01016073 KRT81695.1 NEVH01024531 PNF16976.1 AGBW02009651 OWR50240.1 NNAY01000295 OXU29396.1 GEDC01030992 JAS06306.1 GBHO01021951 JAG21653.1 NWSH01004565 PCG64919.1 AGBW02003464 OWR55546.1 ODYU01000799 SOQ36122.1 GBHO01006039 JAG37565.1 GECL01002483 JAP03641.1 GDHC01019650 JAP98978.1 AJVK01012081 AJVK01012082 GDRN01063798 JAI64926.1 GBHO01006041 GBHO01006040 GBRD01002176 JAG37563.1 JAG37564.1 JAG63645.1 GDHC01003426 JAQ15203.1 GDRN01063797 JAI64927.1 KYM92092.1 GDRN01063799 JAI64925.1 KQ434823 KZC07237.1 DS235156 EEB12660.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000283053
UP000192223
UP000235965
+ More
UP000242457 UP000009046 UP000279307 UP000076502 UP000000311 UP000078541 UP000027135 UP000007266 UP000005203 UP000079169 UP000008237 UP000015103 UP000007755 UP000078492 UP000078540 UP000053825 UP000005205 UP000053105 UP000075809 UP000002358 UP000078542 UP000007819 UP000053097 UP000007151 UP000215335 UP000218220 UP000092462
UP000242457 UP000009046 UP000279307 UP000076502 UP000000311 UP000078541 UP000027135 UP000007266 UP000005203 UP000079169 UP000008237 UP000015103 UP000007755 UP000078492 UP000078540 UP000053825 UP000005205 UP000053105 UP000075809 UP000002358 UP000078542 UP000007819 UP000053097 UP000007151 UP000215335 UP000218220 UP000092462
Interpro
SUPFAM
SSF52091
SSF52091
Gene 3D
ProteinModelPortal
H9JBZ3
A0A194PIW7
A0A0N1PJT0
A0A3S2LU88
A0A1W4XDG5
A0A2J7RSL0
+ More
A0A2A3EE05 A0A1B6MPW8 E0VCB0 A0A3L8DVY2 V9I9F7 A0A154PRJ3 A0A1W4XEP4 E2AWV5 A0A1W4XEN4 A0A1W4XEM9 A0A1B6CML3 A0A1Y1MPJ6 A0A195FJ73 A0A2J7QAB9 A0A2J7QAE0 A0A067RPK8 A0A1B6C3Z6 A0A139WD39 A0A069DWI2 A0A088AR89 A0A2A3EBU0 A0A1B6EVA9 A0A023F3H2 A0A3Q0J7I5 E2BSP6 A0A0P4VQI8 T1IES7 A0A069DV45 A0A023F3Y6 A0A224X9U9 A0A139WCX6 F4WUD0 A0A151J6H4 A0A195BVT9 H9JBZ6 A0A0L7RCE8 A0A151I5C0 A0A158NL57 E2ANG6 A0A154PR24 A0A194PIX2 A0A158NTY8 A0A0M9A911 A0A151X835 A0A195FKC5 A0A2S2NQ78 D2A570 T1HT78 K7IX01 A0A0T6BGS6 K7IYL9 A0A195C7D8 J9JIX9 A0A026X0F7 A0A0C9Q9Z3 A0A2W1BKM4 A0A151I4S0 A0A158NTY7 A0A2J7RSK1 A0A0T6B2L2 A0A2J7PKW2 A0A212F938 A0A232FF55 A0A1B6BYH8 A0A0A9XS76 A0A1W4X674 A0A2A4IZ56 A0A212FP71 A0A2H1V6V8 A0A0A9Z6Y1 A0A0V0G6Q9 A0A146KSN0 A0A1B0D6L8 A0A0P4WEK6 A0A0A9Z2J6 A0A146M619 A0A0P4WSY8 A0A195BU35 A0A0P4W8Z7 A0A154P5U0 E0VH04
A0A2A3EE05 A0A1B6MPW8 E0VCB0 A0A3L8DVY2 V9I9F7 A0A154PRJ3 A0A1W4XEP4 E2AWV5 A0A1W4XEN4 A0A1W4XEM9 A0A1B6CML3 A0A1Y1MPJ6 A0A195FJ73 A0A2J7QAB9 A0A2J7QAE0 A0A067RPK8 A0A1B6C3Z6 A0A139WD39 A0A069DWI2 A0A088AR89 A0A2A3EBU0 A0A1B6EVA9 A0A023F3H2 A0A3Q0J7I5 E2BSP6 A0A0P4VQI8 T1IES7 A0A069DV45 A0A023F3Y6 A0A224X9U9 A0A139WCX6 F4WUD0 A0A151J6H4 A0A195BVT9 H9JBZ6 A0A0L7RCE8 A0A151I5C0 A0A158NL57 E2ANG6 A0A154PR24 A0A194PIX2 A0A158NTY8 A0A0M9A911 A0A151X835 A0A195FKC5 A0A2S2NQ78 D2A570 T1HT78 K7IX01 A0A0T6BGS6 K7IYL9 A0A195C7D8 J9JIX9 A0A026X0F7 A0A0C9Q9Z3 A0A2W1BKM4 A0A151I4S0 A0A158NTY7 A0A2J7RSK1 A0A0T6B2L2 A0A2J7PKW2 A0A212F938 A0A232FF55 A0A1B6BYH8 A0A0A9XS76 A0A1W4X674 A0A2A4IZ56 A0A212FP71 A0A2H1V6V8 A0A0A9Z6Y1 A0A0V0G6Q9 A0A146KSN0 A0A1B0D6L8 A0A0P4WEK6 A0A0A9Z2J6 A0A146M619 A0A0P4WSY8 A0A195BU35 A0A0P4W8Z7 A0A154P5U0 E0VH04
PDB
5DA0
E-value=0.0390989,
Score=88
Ontologies
GO
PANTHER
Topology
Subcellular location
Membrane
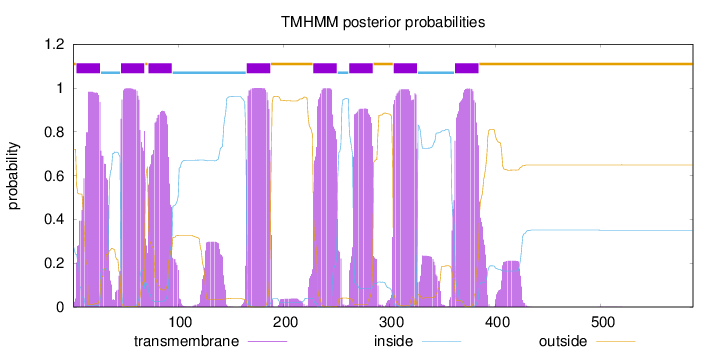
Length:
587
Number of predicted TMHs:
8
Exp number of AAs in TMHs:
191.24938
Exp number, first 60 AAs:
37.99071
Total prob of N-in:
0.27982
POSSIBLE N-term signal
sequence
outside
1 - 3
TMhelix
4 - 26
inside
27 - 45
TMhelix
46 - 68
outside
69 - 71
TMhelix
72 - 94
inside
95 - 164
TMhelix
165 - 187
outside
188 - 227
TMhelix
228 - 250
inside
251 - 261
TMhelix
262 - 284
outside
285 - 303
TMhelix
304 - 326
inside
327 - 361
TMhelix
362 - 384
outside
385 - 587
Population Genetic Test Statistics
Pi
233.539089
Theta
180.770091
Tajima's D
0.757067
CLR
0.346641
CSRT
0.589820508974551
Interpretation
Uncertain
